1. Introduction
The rate of industrial development of cities contributes to the deterioration in the ecological condition of the environment. One of the main pollutants comprises various classes of hydrocarbons—products of crude oil conversion.
Polycyclic aromatic hydrocarbons (PAHs) are formed as a result of various biological processes [1] and are also products of incomplete burning of natural or anthropogenic sources (car exhaust gases, domestic heating, and cigarette smoke) [2]. These substances belong to the group of lipophilic organic pollutants, which are dangerous for the environment and human health [3]. Alkanes are saturated hydrocarbons that can have a linear (n-alkanes), cyclic (cycloalkanes), or branched (isoalkanes) structure. These hydrocarbon molecules are non-polar and chemically inert and have low solubility in water, making them difficult to metabolize by microorganisms. Such molecules have a tendency to accumulate in cell membranes and require additional energy to activate. However, some microorganisms (both aerobic and anaerobic) can use different types of hydrocarbons as the sole source of carbon and energy [4].
Some microorganisms have been found to be capable of degrading both PAHs and alkanes [5,6]. Moreover, the PAH and alkane degradation genes in microorganisms can be localized both on different plasmids [7] and on the same plasmid [8]. A total of 29 monooxygenase and 54 dioxygenase genes were found in the genome of Paraburkholderia aromaticivorans strain BN5. The biodegradation of different hydrocarbons, such as benzene, toluene, ethylbenzene, xylene, and naphthalene, can be linked to similar genes. It should be noted that of the sequences found in the Paraburkholderia aromaticivorans BN5 genome, six monooxygenase and fourteen dioxygenase genes are located on plasmids, and two alkane-1-monooxygenase (alkB) genes have chromosomal localization (CJU94_RS03555 on chromosome 1 and CJU94_RS23135 on chromosome 2) [9]. To date, there is little information on bacterial strains in which genes encoding PAH- and n-alkane-degrading enzymes have been detected simultaneously on the same chromosome [10,11].
Bacteria of the genus Pseudomonas are the most studied microorganisms capable of using hydrocarbons as a sole source of carbon and energy and producing bioactive substances that contribute to the absorption of such compounds. Pseudomonas are often used to study the biodegradation of various biogenic and xenobiotic pollutants. This is due to their metabolic universality and participation in various environmental processes.
Genomes of Pseudomonas strains belonging to different species have been characterized, e.g., P. entomophila (e.g., strain L48), P. putida (e.g., strains GB1, KT2440, F1, W619, S16, CSV86, SF1, DOT-T1E, B6-2, DLL-E4), P. stutzeri (e.g., strains KF716, A1501, CCUG29243), P. protegens (strain Pf-5), P. fluorescens (strains Pf-01, SBW25), P. knackmussi (strain B13), P. aeruginosa (e.g., strains PA7, PAO1, PACS2, 2192, and PA14), P. pseudoalcaligenes (strain KF707), P. azelaica (strains Aramco J, HBP1), P. syringae (strains B728a, DC3000 and 1448A,), and P. mendocina (strain YMP). The size of Pseudomonas genomes is about 6 Mb [12], and many genes are involved in the catabolism of different carbon sources and their adaptation to exist in a specific ecological niche.
In most cases, aromatic catabolism genes are organized into gene clusters that also contain specialized regulatory and transport genes and, in some cases, efflux pump genes [13]. For example, the naphthalene degradation genes, often used as a model substrate to study the various aspects of PAH catabolism, are organized into two operons in pseudomonads. Nine genes (nahAaAbAcAdBFCED) form the upper pathway operon (nah operon), which catalyzes the conversion of naphthalene to salicylate. The other 10 genes (nahGTHINLOMKJ) comprise the lower pathway operon (sal operon), which encodes the genes for the conversion of salicylate to tricarboxylic acid cycle intermediates by the enzymes of the catechol meta-cleavage pathway [8]. The P. putida AK5 strain is also known to contain the genes (sgpGHIKGHB) for the conversion of salicylate to tricarboxylic acid cycle intermediates via enzymes of the gentisate cleavage pathway [14]. Despite the fact that most naphthalene degradation genes identified in different bacteria have 99–100% identity with their analogs in other strains, the localization (plasmid or chromosome) and organization of these gene clusters may differ in each strain [15].
In the octane (OCT) plasmid of Pseudomonas putida strain GPo1, the alkane-degrading genes are organized as the alkBFGHJKL operon and are controlled by the products of another operon (alkST) located 40 bp downstream of the first operon [14]. van Beilen, J. B et al. [16] described a strain of P. putida P1 that contains the same operon structure (alkBFGHJKL), but in which alkST has been moved upstream of the operon and the alkL and alkN genes are not separated by the insertion sequence (IS) [15].
It is known that Pseudomonas aeruginosa strain PAO1 is capable of degrading n-alkanes, naphthalene, and phenanthrene [10,11,17,18]. Furthermore, the genes responsible for the degradation of the above substrates are located on the chromosome. The work of Qi, J. et al. [10] demonstrated the presence of functional genes of the naphthalene degradation pathway by gentisate in the PAO1 strain genome. Theo H.M. Smits et al. [11] demonstrated the presence of two alkane monooxygenase genes (alkB1 and alkB2) homologous to the alkane monooxygenase gene (alkB) of strain P1. The same authors also showed the presence of functional rubredoxin reductase genes (rubA1, rubA2, and rub). Later [19], a cluster of lao genes was discovered, presumably involved in the degradation of primary long-chain alcohols. However, no detailed description of the n-alkane degradation pathway in Pseudomonas aeruginosa strain PAO1 has been found in the literature.
The process of microbial biodegradation of hydrocarbons involves adhesion to the substrate and the formation of biosurfactants/bioemulsifiers, biopolymers, solvents (e.g., acetone, ether, benzene), gases, and acids (e.g., stearic acid) to increase the bioavailability of hydrocarbon substrates. These substances also contribute to substrate detoxification [3,5,20].
The presence of oil hydrocarbons in the environment affects bacterial survival. Destruction of such substances often requires microbial adaptation to the pollutant, which is an important strategy for increasing the stress resistance and tolerance of microbial cells. Oil hydrocarbons, being toxic and persistent, act as a driver of natural selection in the microbial community, contributing to the evolution of degradation pathways. One example of microbial adaptation to extreme environmental conditions is the formation of biofilm [21]. A biofilm is an association of bacteria containing a slimy extracellular matrix with cells that can bind to hydrocarbons and enhance solubilization, leading to the degradation of these substrates. In addition, the hydrophobic nature of the bacterial surface has a direct effect on biofilm formation. Such an adaptation factor allows microorganisms to bind to surfaces of biotic and abiotic origin [22].
Despite the fact that many researchers have studied microorganisms that degrade oil hydrocarbons, very little work has been undertaken on the genomics and physiology of microorganisms capable of simultaneously degrading PAHs and n-alkanes. Pseudomonas aeruginosa PAO1, a multi-destructor of various hydrocarbons, has been the most extensively studied. However, this bacterium is pathogenic, making its use in environmental remediation technologies impossible. However, such multi-degrading strains can be used as model systems to study the mechanisms of simultaneous degradation of different classes of hydrocarbons. This is certainly important for the development and improvement of technologies for the bioremediation of oil-contaminated sites.
The aim of the present study was to investigate the genetic and physiological characteristics of the Pseudomonas sp. strain OVF7 isolated from oil-contaminated soils of the “Pogranichnoye” oil field in Yamalo-Nenets Autonomous Okrug (Russia), which is capable of degrading both PAHs and n-alkanes and forming biofilms when grown in liquid media with carbon substrates.
A probably new bacterial species of the group of fluorescent pseudomonads, capable of forming biofilms having different architectures in the degradation of naphthalene, n-dodecane, and a mixture of these hydrocarbons, was isolated and is described herein. The simultaneous presence on the chromosome of gene clusters of the naphthalene degradation pathway via salicylate using the meta-pathway of catechol cleavage and gene clusters of the n-alkane degradation pathway via monoterminal oxidation was found. The genes encoding enzymes of the “classical” complete pathways of degradation of n-alkanes and naphthalene to Krebs cycle intermediates were described in the strain under study. A strain of Pseudomonas sp. strain OVF7 is interesting both as a model object for studying the mechanisms of degradation of different classes of hydrocarbons and as a promising agent for application in environmental clean-up biotechnologies.
2. Materials and Methods
2.1. Bacterial Strain
The strain OVF7 was isolated from the oil-contaminated soils of the “Pogranichnoye” oil field in the Yamalo-Nenets Autonomous Okrug (Russia). Strain OVF7 was stored in the collection of the Laboratory of Plasmid Biology (IBPM RAS (FRC PSCBR RAS), Pushchino, Russia). It is capable of utilizing n-alkanes, naphthalene, crude oil, and diesel fuel at 28 °C.
2.2. Chemicals
High purity (>98%) analytical-grade sodium succinate, dichloromethane, naphthalene, n-dodecane, n-octane, n-nonane, n-undecane, n-hexadecane, and n-decane were purchased from Sigma-Aldrich (St. Louis, MO, USA).
2.3. Genome Sequencing, Assembly and Annotation
A cetyltrimethylammonium bromide miniprep procedure [23] was used to isolate and purify total DNA from Pseudomonas sp. strain OVF7. Illumina and Oxford Nanopore technologies were used for DNA sequencing of Pseudomonas sp. strain OVF7. The FLAMING 106 flow cell (Oxford Nanopore Technologies (Oxford, UK) [ANT]) was used for sequencing on the MinION equipment. Using the ligation sequencing kit (catalog number SQK-L SK 109), libraries were prepared. In parallel, the S2 reagent kit (catalogue number 20012861; 2 × 100 bp) was used for nucleotide sequencing on the Illumina NovaSeq 6000 platform. The paired-end library was generated using the KAPPA Hyper Plus Kit (KAPA biosystems, Wilmington, MA, USA).
The programs SPAdes version 3.15.2 [24], Unicycler (May 2023) [25], and Flye 2.9 [26] were used to assemble the hybrid based on the Nanopore and Illumina reads. We used Pilon version 1.23 [27] and Bowtie2 version 2.3.5.1 [28] for error correction. To confirm that we had assembled a circular replicon, we analyzed the presence of overlapping ends. To make the results public, the data were deposited in the GenBank (BioProject, PRJNA987530; BioSample, SAMN35982525; GenBank, NZ_ CP128996).
Prokka software v1.14.5 helped us to annotate the genome [29]. We used the Basic Local Alignment Search Tool (BLAST) [30] to search for functions of some hypothetical proteins.
Taxonomic analysis of the complete genome assembly was performed on the Type (Strain) Genome Server (TYGS) (https://tygs.dsmz.de (accepted date (2 June 2023)) [31], taking into account recently developed methodological updates [32]. The MASH algorithm [33] was used to search for genomes of closely related strains in comparison with the genome of Pseudomonas sp. OVF7. In addition, the 16S rDNA gene sequence was used to select 10 closely related type strains. Using RNAmmer [34] and BLAST [30], the 16S rDNA gene sequences of 19,149 type strains available in the TYGS database were extracted and compared with the same sequence in the genome of Pseudomonas sp. strain OVF7. According to the algorithm [35], the top 50 matching type strains were selected (according to the bit ratio). A final selection of the 10 closest type strain genomes was made using the distance formula d5 [35]. Digital DNA–DNA hybridization (DDH) values and confidence intervals were calculated using the recommended settings of GGDC 3.0 [32,36].
FASTME 2.1.6.1 [37] was used to construct a minimum evolutionary tree based on the intergenomic distances obtained. Tree rooting was performed at the midpoint [38] and visualized using https://itol.embl.de/ (accepted date (9 June 2023)) [39]. Species and subspecies were clustered according to [31] and [40], respectively. The OrthoANI algorithm [41] was used to calculate the average nucleotide genome identity (ANI) between strains of Pseudomonas sp. strain OVF7 and related strains.
The rpoD gene phylogenetic tree was built using the neighbor-joining method (BV-BRC) [42], accessed 25 May 2023. The CGView program [43] was used to image the chromosome map of Pseudomonas sp. strain OVF7.
2.4. Growth Media and Conditions
To prepare the inoculum, the cell culture was grown in liquid mineral medium containing 2% (v/v) succinate. The suspension obtained was centrifuged and the sediment was washed with physiological solution. The sediment was then resuspended in physiological solution and the cell concentration in the suspension was increased to 1 × 108 CFU/mL using a turbidity standard. The inoculum was added to the test systems until the final concentration was about 1 × 106 CFU/mL.
The Pseudomonas sp. strain OVF7 was grown in Erlenmeyer flasks containing 100 mL of mineral medium with the following composition [44]. The flasks were sealed with sterile rubber stoppers to prevent abiotic losses of the substrates tested. The plugs were opened in a sterile manner for 2–3 min daily to renew the gas phase, taking into account the amount of oxygen in the system. Succinate, naphthalene, phenanthrene, toluene, benzene, anthracene, n-octane, n-nonane, n-dodecane, n-undecane, n-dodecane, and n-hexadecane were used as carbon and energy sources to assess the substrate specificity of the strains. Substrates were added at 1 g/L. The strain was grown in flasks for 1–7 days at 28 °C and 180 rpm. Visual assessment of culture growth was performed throughout the period.
To plot the growth curve, a suspension was taken and diluted ten-fold, and then the suspension was seeded onto Lysogeny Broth (LB) agar medium [45]. Plates were incubated at 28 °C for 2 days. Individual colonies were then counted. Growth curves were constructed from the data obtained.
The loss of naphthalene, n-dodecane, and the mixture of naphthalene and n-dodecane was evaluated in the described systems at the end of the exponential growth phase. For the evaluation of abiotic losses of introduced hydrocarbons, systems without microorganisms were used for the same period of time. All results were derived from five independent replicates.
At the end of the exponential growth phase, Pseudomonas sp. strain OVF7 was also sampled for further microscopic studies. For microscopy, the culture grown in mineral medium with succinate as the sole source of carbon and energy at the end of the exponential growth phase was used as a control system.
2.5. Determination of Hydrocarbon Content in the Medium
Dichloromethane (1:1, v/v) was used to extract n-dodecane and naphthalene from the growth medium. Substrate content was estimated by gas chromatography (Agilent 6890, Agilent Technologies, Santa Clara, CA, USA) using a flame ionization detector. The chromatographic column was DB-1 (30 m × 0.25 mm id, 0.25 µm). The oven temperature was increased by 15 °C every minute (the initial temperature was 40 °C). The Collaborative Use Center, Department of Soil Science, and Lomonosov Moscow State University kindly provided equipment for sample analysis. Analytical standards were used for absolute calibration and quantification. The coefficient of correlation amounted to 0.99. ANOVA was p = 0.04. Samples were diluted 100-fold prior to analysis.
The degree of hydrocarbon biodegradation (D) was calculated using the following formula:
2.6. Light and Fluorescent Microscopy
Cells were examined by phase-contrast and fluorescent microscopy in an AXIO Imager A1 (Zeiss, Oberkochen, Germany) with a filter set 56HE (Zeiss) at a wavelength of 490 nm (excitation) and 512 + 630 (emission). An Axiocam 506 (Zeiss) was used for image acquisition. Live and dead cells were revealed using a LIVE/DEAD™ BacLight™ Bacterial Viability Kit (Molecular Probes, Eugene, OR, USA).
2.7. Scanning Electron Microscopy
The surface morphology of the biofilms was examined using scanning electron microscopy (SEM). Samples of the cells placed on membrane filters were fixed in glutaraldehyde vapor for 24 h at 4 °C and post fixed in OsO4 vapor for 3 h at 20 °C. After dehydration in propylene oxide vapor, the samples were coated with gold (Fine Coat Ion Sputter JFC-1100, Tokyo, Japan) and examined under a scanning microscope JSM-6510LV (JEOL, Tokyo, Japan).
3. Results
3.1. Nucleotide Sequence and Annotation
Sequencing and complete assembly of the genome of Pseudomonas sp. strain OVF7 revealed a circular chromosomal replicon of 7,174,656 bp (GC content: 60.35%). The chromosome contains 6628 coding sequences (CDS), 6 rRNAs, and 62 tRNAs. A function was assigned to 5147 CDS, of which 1480 CDS were annotated as hypothetical proteins. We identified genes involved in naphthalene and n-alkane degradation. We also found several genes for incomplete degradation pathways for caprolactam, toluene, biphenyl, xylene, benzoate, styrene, 2,4-dichlorobenzoate, atrazine, ethylbenzene, anthracene, biphenyl, and fluorobenzoate.
On the basis of the preliminary analysis of the 16S rRNA gene, the studied strain was assigned to the genus Pseudomonas, and specifically to the group of fluorescent pseudomonads. Based on the whole-genome sequencing, an attempt was made to determine the phylogenetic relationship between the studied strain and its closest type strains (Figure 1A). However, the whole-genome alignment did not allow an accurate determination of species affiliation. In accordance with the recommendations of Chun et al. [46], the Average Nucleotide Identity and digital DNA–DNA Hybridization values were used to compare the genome of Pseudomonas sp. strain OVF7 genome with those of closely related type strains. The comparisons of ANI and DDH for the strain Pseudomonas sp. strain OVF7 with these values for the strains of species of the group of fluorescent pseudomonads listed in were 84.44–88.87% and 36.6–59.1%, respectively. Articles [36,46,47] suggest that when describing new species, these indices should be lower than 95% (ANI) and 70% (DDH) in comparison with type strains of known species.

Figure 1. Phylogenetic trees of 49 type strains of Pseudomonas and the study strain OVF7. (A) Phylogenetic tree of the whole and draft genomes of the Pseudomonas type strains and Pseudomonas sp. OVF7. (B) Phylogenetic tree based on the rpoD gene. Maximum likelihood tree, GTR + G + I model (MEGA-X)) including, respectively, 49 type strains of Pseudomonas and the study strain, respectively. Bootstrap values were calculated on the basis of 1000 replications. The Pseudomonas sp. strain OVF7 is indicated in red font.
According to the article by Girard L. et al. [48], the group of fluorescent pseudomonads includes the following species: P. allii, P. antartica, P. asgharzadehiana, P. aylmerense, P. azadiae, P. azotoformans, P. canadensis, P. carnis, P. cedrina subsp. cedrina, P. cedrina subsp. fulgida, P. costantinii, P. cremoris, P. cyclaminis, P. edaphica, P. extremaustralis, P. carnis. cremoris, P. cyclaminis, P. edaphica, P. extremaustralis, P. extremorientalis, P. fildesensis, P. fluorescens, P. grimontii, P. haemolytica, P. kairouanesis, P. karstica, P. khavaziana, P. kitaguniensis, P. lactis, P. libanensis, P. lurida, P. marginalis, P. nabeulensis, P. orientalis, P. palleroniana, P. panacis, P. paracarnis, P. paralactis, P. paralactis, P. pisciculturae, P. poae, P. rhodesiae, P. salmasensis, P. salomonii, P. simiae, P. sivasensis, P. spelaei, P. synxantha, P. tolaasii, P. tritici, P. trivialis, P. veronii, P. yamanorum. The authors recommended the estimation of the phylogenetic relationships of the strains on the basis of the rpoD gene (Figure 1B). However, even using this approach, it was not possible to determine the species assignment of the strain. It can be noted that, based on the trees presented (Figure 1A,B), the closest species to the studied strain are P. veronii and P. fildesensis. However, there are currently no whole-genome type strains of these two species. So, it is not possible to make a detailed comparison of genome structure using, for example, the Mauve method.
Thus, based on the combination of approaches used (phylogenetic tree of the whole/draft genomes of the Pseudomonas type strains, phylogenetic tree based on the rpoD gene, ANI and DDH indexes), the isolated microorganism can be classified as a new species of the group of fluorescent pseudomonads.
3.2. Genes Potentially Involved in n-Alkanes and Naphthalene Degradation
Analysis of the whole-genome sequence of strain OVF7 showed that its chromosome contains all the genes that, according to literature data [16], are necessary for the conversion of n-alkanes to acyl-CoA derivatives. These genes are located in a chromosome region of about 15 kb and are organized into the two “classical” operons, alkST and alkBFGHJKL. The mutual arrangement of the n-alkane oxidation gene clusters and the overall organization of this chromosome region in strain OVF7 are identical to those in the strains P. veronii strain 7–41 (CP089552.1) and P. putida strain P1 (GCA_001865225.1) [8,14]. Note that in the case of P. veronii strain 7–41, the alk-genes are plasmid localized, whereas in the P. putida strain P1, the genes are localized within the chromosome. In addition, the n-alkane degradation genes in the strain under study, as in the cases of P. putida strain P1 and P. veronii strain 7–41, are flanked by identical copies of the insertion sequence ISPpu4. In general, the region containing the alk-genes clusters and flanking their mobile elements occupies a region of about 23 kb.
The identity of the nucleotide and deduced amino acid sequences of the n-alkane oxidation genes with the corresponding sequences available in databases was analyzed using the BLAST online resource (accessed on 20 June 2023). These indices were 100% for almost all alk-genes of strain OVF7 with the corresponding sequences of the strains P. veronii strain VI4T1 (ASM202932v1), P. veronii strain 7–41 (pPCP7-41), and P. putida strain P1 . The exception was the gene encoding long-chain-fatty-acid-CoA ligase, alkK, for which the amino acid sequence identity was 99%. The organization of the n-alkane degradation genes was studied in detail using the OCT plasmid of the P. putida strain GPo1 as an example. The identities of the deduced amino acid sequences of the alk-genes of the OVF7 strain with the corresponding sequences of the OCT plasmid ranged from 47% to 92%. Moreover, the highest value was observed for alkane 1-monooxygenase, AlkB, and the lowest for rubredoxin-1, AlkF .
In the chromosome of strain OVF7, the naphthalene and salicylate degradation gene clusters (nah and sal genes) are located downstream of the alkBFGHJKL gene cluster. The distance between the alk and sal genes is about 7 kb. The genetic organization of the naphthalene biodegradation system in pseudomonads has been well studied in the P. putida strain G7 carrying the NAH7 plasmid [49,50,51]. All genes required for the conversion of naphthalene to Krebs cycle intermediates in the P. putida strain G7 have plasmid localization. It should be noted that with the development of sequencing technologies it has been found that in pseudomonads the nah genes have about 90% homology with the corresponding genes of the NAH7 plasmid [8,52,53]. To determine the degree of similarity between the nucleotide and deduced amino acid sequences of the naphthalene degradation genes of strain OVF7, we compared the nah gene clusters with the corresponding sequences of other bacteria, including pseudomonads, deposited in public databases. In strain OVF7, as in the case of plasmids pCP7-41 (CP089552.1), pND6-1 (AY208917.2), pDTG1 (AF491307.2), and NAH7 (NC_007926.1), the naphthalene degradation genes are organized into two operons: the nah operon and the sal operon. The nah operon contains nine genes (nahAaAbAcAdBFCED) encoding enzymes for the oxidation of naphthalene to salicylate. The sal operon includes genes (nahGTHINLOMKJ) necessary for the cleavage of salicylate to Krebs cycle intermediates. Unlike the above plasmids, the nah and sal operons of OVF7 are a chromosomally localization. The genes belonging to the nah operon occupy a chromosome region of 9 kb, whereas the size of the sal operon is 10 kb. It should also be noted that the nah and sal genes are transcribed in opposite directions from each other. The identities of the deduced amino acid sequences of the nah and sal operon genes of strain OVF7 with the corresponding sequences of the naphthalene catabolic plasmids NAH7 (NC_007926. 1), pND6-1 (AY208917.2), pDTG1 (AF491307.2), pCP7-41 (CP089552.1), and the unnamed plasmid of strain P. veronii strain Pvy (CP039632.3), were 72–100% . The lowest similarity was shown with the catabolic genes of plasmid NAH7 and the highest similarity with pCP7-41. The identity of the deduced amino acid sequences of almost all nah and sal genes between OVF7 and pCP7-41 was 100%, except for the gene encoding the outer membrane beta-barrel protein (nahQ). It was 99% for NahQ. Clusters of nah and sal genes were also found in P. veronii strain VI4T1. The identity of the deduced amino acid sequences of the genes responsible for naphthalene catabolism in P. veronii strain VI4T1 and Pseudomonas sp. strain OVF7 was 100% for all genes. A CDS with a high degree of homology of the deduced amino acid sequence with the corresponding sequence of plasmid pCP7-41 (UHH01036.1) and the unnamed plasmid of P. veronii strain Pvy (CP039632.3) was found downstream of the sal operon (100% and 95%, respectively). This CDS encodes a LysR-type transcriptional regulator required for the expression of the nah and sal operons. The IS elements of the IS110 family transposase flank the nah operon of strain OVF7.
The mutual arrangement of the n-alkane and naphthalene degradation genes in the OVF7 strain chromosome is such that the nah genes are located downstream of the sal genes, while the alk genes are located upstream of the sal genes. The genetic organization of the alk–sal–nah region in the Pseudomonas sp. strain OVF7 chromosome is completely consistent with that of the P. veronii strain 7–41 (pCP7-41 plasmid) (Figure 2). In addition, the identity of the nucleotide sequences of these regions in both strains was 99%.
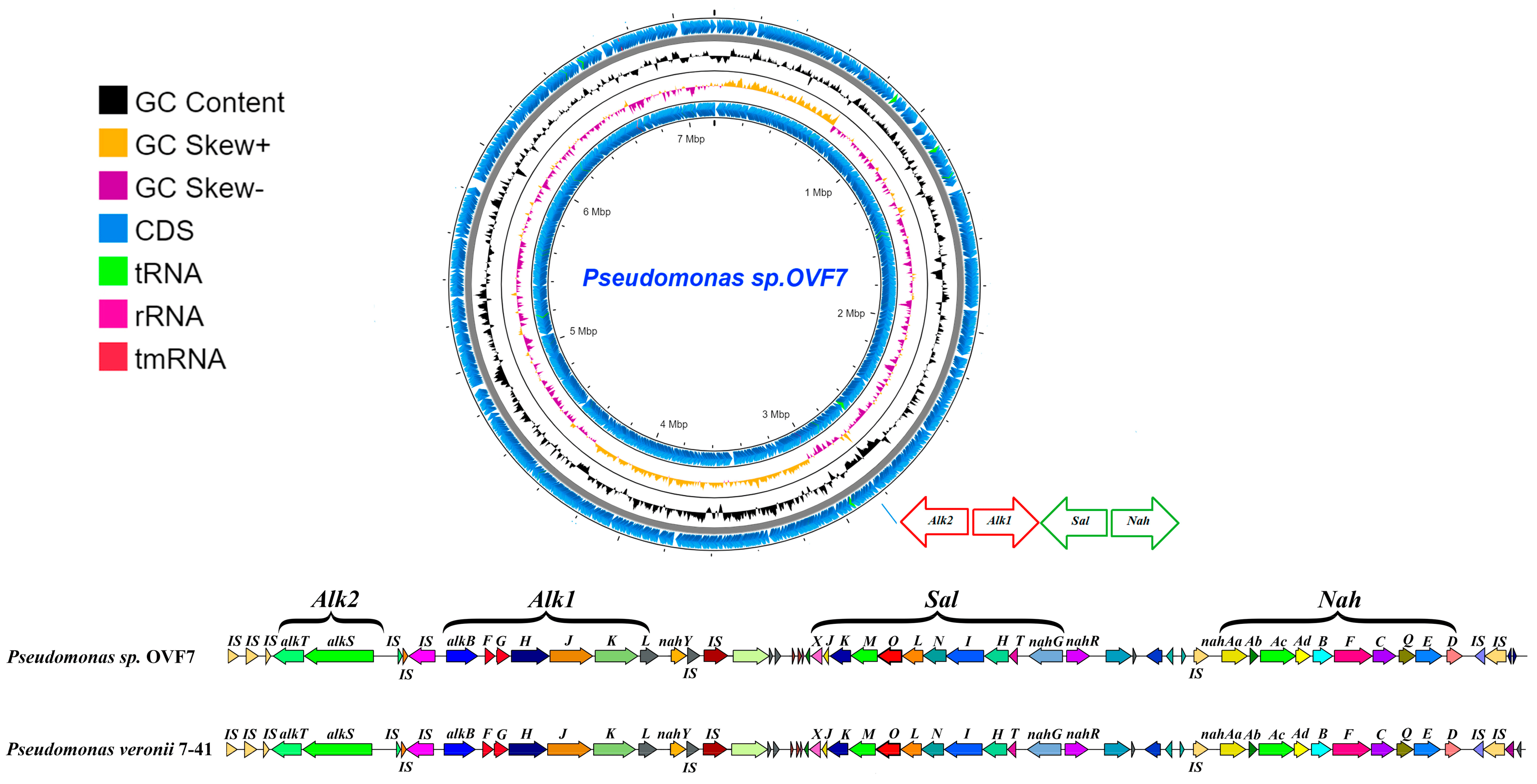
Figure 2. Circular map of the Pseudomonas sp. OVF7 chromosome and comparison of the alk–sal–nah region in strains Pseudomonas sp. OVF7 and P. veronii 7-41 (pCP7-41 plasmid). From outside to the center: all CDS and RNA genes on forward strand, GC skew, GC content, all CDS and RNA genes on reverse strand.
3.3. Growth of Pseudomonas sp. OVF7 in Mineral Medium with n-Dodecane and Naphthalene
The ability of Pseudomonas sp. strain OVF7 to grow at 28 °C in liquid mineral medium on different PAHs and n-alkanes was studied. As a result, the strain did not grow on the following substrates: phenanthrene, toluene, benzene, anthracene, and n-hexadecane. The strain OVF7 showed high activity in the medium containing naphthalene and during growth on n-alkanes (n-octane, n-nonane, n-decane, n-dodecane, and n-undecane). The formation of small flakes was observed when the strain grew in the culture medium containing n-octane, n-nonane, n-decane, and n-undecane. However, in the medium with n-dodecane as the sole source of carbon and energy, the formation of a dense layer of small bubbles was observed at the culture medium–hydrocarbon interface, while no flakes were observed in the remaining part of the medium. It was decided to use n-dodecane as a model n-alkane for further work. Strain OVF7 was also able to utilize a mixture of naphthalene and n-dodecane in a liquid mineral medium. To assess the abiotic loss of substrates (naphthalene and/or n-dodecane), an experiment was carried out in flasks containing hydrocarbons without microorganisms. Abiotic loss was practically not observed and was about 1–2% over 7 days because the flasks were sealed with rubber plugs.
Growth curves of Pseudomonas sp. strain OVF7 obtained by culturing the strain in a liquid mineral medium containing naphthalene (1 g/L), n-dodecane (1 g/L), and a mixture (naphthalene—n-dodecane—1 g/L (0.5 g/L each substrate)) were constructed (Figure 3).
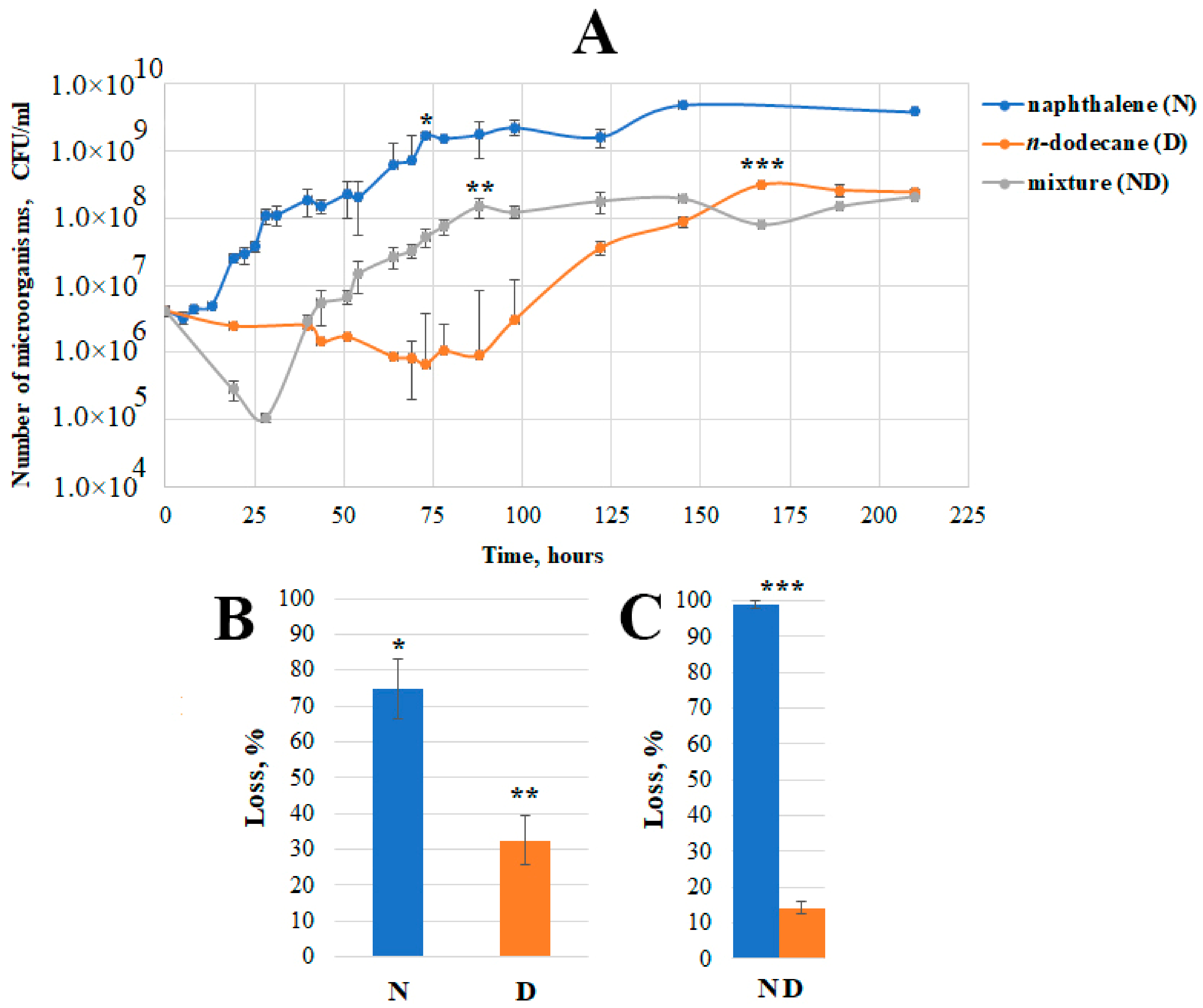
Figure 3. Physiological parameters of growing Pseudomonas sp. strain OVF7 in mineral medium with naphthalene (N), n-dodecane (D), and a mixture of naphthalene and n-dodecane (ND). (A)—Growth curves; (B)—Loss of naphthalene and n-dodecane in systems containing only one of these substrates; (C)—Loss of naphthalene and n-dodecane in a system containing a mixture of these substrates; *, **, *** on A denote the time of the end of the exponential growth phase corresponding to the naphthalene and/or n-dodecane loss data on (B,C).
As can be seen from Figure 3, when grown in the medium containing naphthalene at 28 °C, the lag phase of the culture was quite short (compared to the lag phase in the n-dodecane or a mixture of naphthalene and n-dodecane) and lasted until about 13 h. A small plateau was observed in the middle of the exponential growth phase (after 30 h of cultivation). This is probably related to the time of adaptation of the strain to the accumulated salicylate (intermediate of the naphthalene degradation pathway) and the beginning of its consumption. The accumulation of salicylate at this time was not investigated in the present work. However, it was previously shown [8] that in the middle of the exponential growth phase on naphthalene in pseudomonads there is a slight accumulation of salicylate and then its consumption. Such a phenomenon is necessary for the activation of sal operon genes. After 75 h of cultivation, the bacterium reached the stationary phase of growth and the number of microorganisms was about 109 CFU/mL, which remained practically unchanged thereafter. At this time, the loss of naphthalene was 75%, i.e., the strain was metabolizing about 0.75 g of the substrate.
A long period of adaptation of the bacterial culture to the substrate (87 h) was observed when growing on n-dodecane (initial concentration 1 g/L) in liquid mineral medium at 28 °C. The exponential phase was monotonous and the culture reached its maximum abundance (108 CFU/mL) at the end of the exponent after 167 h of cultivation. At this time, the loss of n-dodecane was 32%, i.e., the strain metabolized about 0.32 g of the substrate. It is likely that the lower consumption of n-dodecane by the strain influenced the lower maximum cell number compared to that when cultured on naphthalene.
A culture of Pseudomonas sp. strain OVF7 was also grown on a mixture of naphthalene and n-dodecane (with initial substance concentrations of 0.5 g/L naphthalene and n-dodecane, respectively) in liquid mineral medium at 28 °C. As shown in Figure 3, a lag phase was observed during the first 25 h of cultivation, which was accompanied by cell death and a decrease in number by more than an order of quantity. The subsequent exponential phase was similar in character to the growth on naphthalene. The secondary growth (after 50 h) began after a short period characteristic of salicylate accumulation (a similar phenomenon was observed in strain 7–41 when grown on a mixture of PAHs and n-alkane [8]). The stationary phase of growth started a little later than when growing on naphthalene as the sole of carbon and energy source (after 87 h). The maximum cell number was comparable to the maximum number when growing on n-dodecane alone (108 CFU/mL). The degradation rate of naphthalene reached the value of 100% and that of n-dodecane was 12% (Figure 3), which was about 0.56 g/L of carbon substrates.
It can be concluded that the mixture of hydrocarbons (naphthalene–n-dodecane) had a toxic effect on the bacterial culture during the first hours of cultivation of Pseudomonas sp. OVF7. After the adaptation period, the character of the growth curve was as close as possible to the growth curve obtained when the strain was cultured on naphthalene alone.
3.4. Microscopic Data
In addition, the cell precipitate was surrounded by a slime-like substance characteristic of the biofilm matrix. To confirm the ability to form biofilms during growth on different substrates, the Pseudomonas sp. strain OVF7 was cultured in liquid mineral medium containing succinate (as a control), naphthalene, n-dodecane, and a mixture of naphthalene and n-dodecane. The cultured cells were examined by phase-contrast, fluorescence, and scanning electron microscopy.
As shown in Figure 4, Pseudomonas sp. strain OVF7 formed a biofilm. Moreover, the architecture of the biofilm depended on the substrate (Figure 4(A3,B3,C3,D3)). When cultured in liquid mineral medium with succinate, the bacterial cells lined up side by side, forming a dense homogeneous layer (Figure 4(A3)). The strain formed a biofilm whose matrix was perforated by a system of microchannels when naphthalene was used as the sole source of carbon and energy (Figure 4(B3)). The biofilm had a porous spongy structure when the Pseudomonas sp. strain OVF7 was in a bi-substrate system (naphthalene and n-dodecane) (Figure 4(C3)). The use of n-dodecane as the sole substrate resulted in the formation of a dense biofilm layer (Figure 4(D3)).
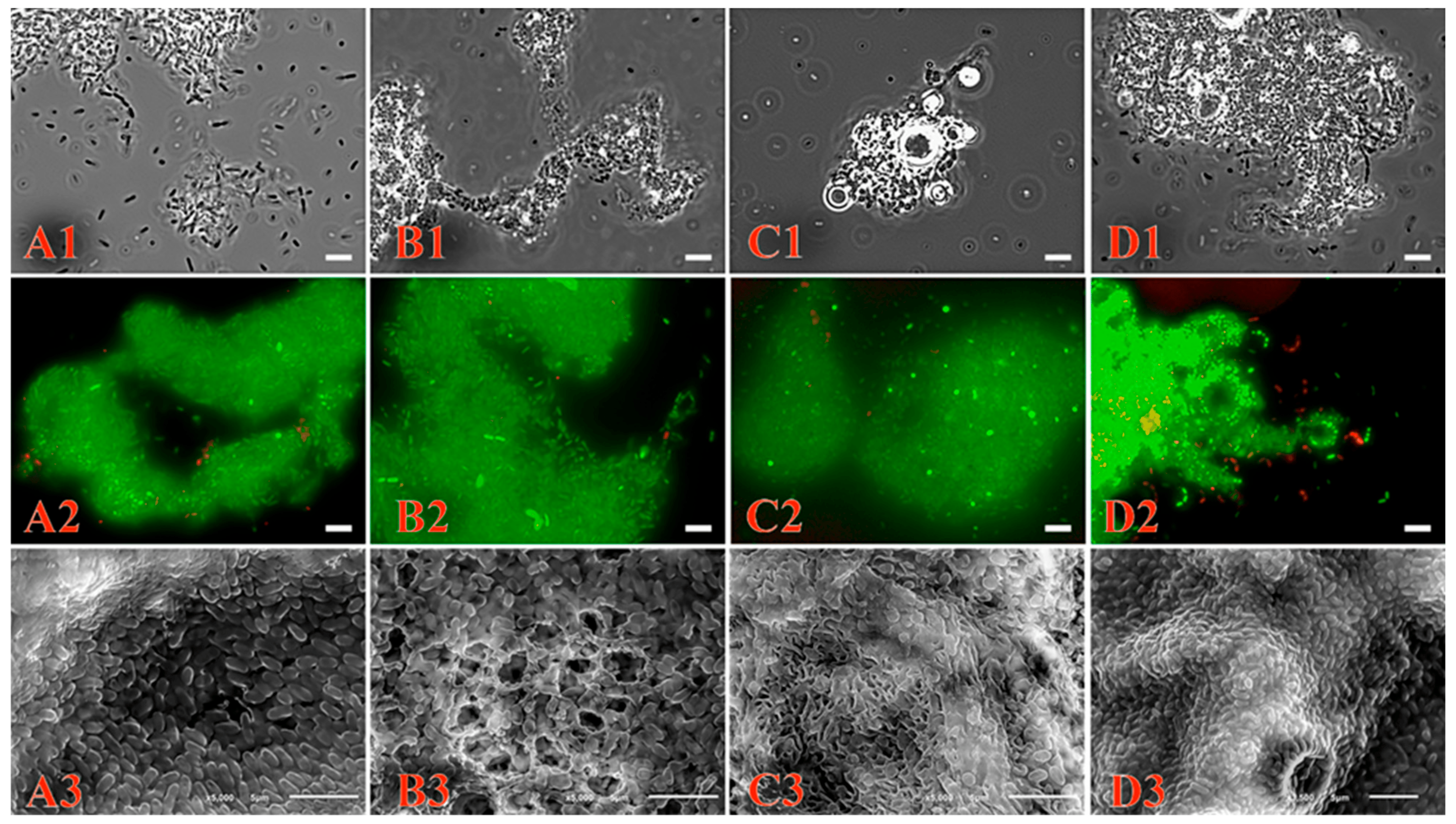
Figure 4. Biofilm formation of Pseudomonas sp. OVF7 when grown in liquid mineral medium with succinate (A1–A3), naphthalene (B1–B3), naphthalene–n-dodecane mixture (C1–C3) and n-dodecane (D1–D3), scale bar—5 µm. (A1,B1,C1,D1)—images obtained by phase-contrast microscopy; (A2,B2,C2,D2)—images obtained by fluorescence microscopy; (A3,B3,C3,D3)—images obtained by scanning electron microscopy.
In addition, images obtained by fluorescence microscopy (Figure 4(A2,B2,C2,D2)) showed the formation of biofilms from living cells.
4. Discussion
Pseudomonas has been actively studied since 1894. These strains are among the most common genera in different microbial communities. Therefore, these microorganisms are responsible for various ecological functions. The ability of Pseudomonas to degrade various hazardous organic pollutants is well known [54]. Such metabolism is due to both the plasticity of the microbial genome and the presence of horizontal gene transfer as one of the means of bacterial adaptation to anthropogenic environmental pollutants. A better understanding of the genomic architecture and dynamics of bacterial genomes is necessary for the successful development of strategies for bioremediation of contaminated sites and water areas.
Pseudomonas have a large number of assemblies represented in various databases such as NCBI, BV-BRS, TYG, and Pseudomonas.com [31,42]. A detailed taxonomy of this genus is still lacking and, despite its long history of study and extensive databases, is constantly being updated with new results. Analysis of the 16S rRNA gene currently allows only the genus identity of Pseudomonas to be determined with certainty. The modern development of whole-genome sequencing technologies has greatly expanded the base of publicly available genomes. The analysis of genomes, or more precisely, their comparison using the ANI and DDH indices [36,54,55,56,57], has allowed a better identification of bacterial taxa. In our case, a whole-genome sequence (Figure 1A) analysis of the above-mentioned indices of the Pseudomonas sp. strain OVF7 in comparison with type representatives of the genus Pseudomonas did not allow its species identity to be determined. Based on the TYG server data, the strain was only identified as belonging to the group of fluorescent pseudomonads. Girard L. et al. [48] performed a large-scale study of the phylogeny of different Pseudomonas species based on the rpoD gene and whole genome sequences. The authors proposed to use the comparison of rpoD gene sequences as one of the representative approaches for species classification of Pseudomonas, including the group of fluorescent pseudomonads. Based on the rpoD gene (Figure 1B), it was also not possible to determine the species identity of Pseudomonas sp. strain OVF7. Only the closely related species P. veronii and P. fildesensis were detected. Moreover, the synergism of the presented approaches allows us to assume that the strain studied in the present work is a member of a new species of Pseudomonas. Therefore, our future plans include a detailed polyphasic taxonomy using phenotypic, chemotaxonomic, genetic, and phylogenetic methods to confirm or refute this hypothesis.
Analysis of the chromosome of Pseudomonas sp. strain OVF7 revealed the presence of clusters of genes encoding enzymes for the biodegradation of n-alkanes and PAHs (Figure 2). The nucleotide and amino acid sequence similarity of these genes to similar sequences described for destructor strains of the genera Pseudomonas should be noted. In addition, the presence of IS elements at the ends of degradative operons indicates a possible origin of catabolic genes in the genome of the strain under study by horizontal gene transfer from a strain with a similar organization of xenobiotic biodegradation pathways. The presence of IS elements may indicate that the strain under study has potential for application in various technologies of environmental remediation of crude oil and oil products.
Based on analyses of the OVF7 strain genome and data from literature studies, we inferred the pathways of naphthalene and n-dodecane metabolism, which are shown in , respectively. Potential genes encoding degradation enzymes for these compounds are presented in . The multi-degrader Pseudomonas aeruginosa strain PAO1 is currently being studied in detail. The genome of Pseudomonas sp. strain OVF7 contains genes for naphthalene catabolism via the meta-pathway of catechol cleavage to Krebs cycle intermediates. Strain PAO1 also has a complete pathway for naphthalene degradation, but via gentisate. The ability of strain PAO1 to degrade n-alkanes is also known; several homologues of genes responsible for the initial steps of the degradation pathway (monooxygenases and rubredoxins) have been found, but further steps of degradation of the formed metabolites are still under investigation. For example, it is not yet clear whether there is a complete degradation pathway from n-alkanes to Krebs cycle intermediates in strain PAO1. In the studied strain OVF7, the structure of the genes responsible for the degradation of aliphatic compounds is organized into two “classical” operons of the complete pathway of degradation of n-alkanes to Krebs cycle intermediates (a similar structure was found in the OCT plasmid). Since strain PAO1 belongs to pathogenic species, it can only be considered as a model for the study of degradation mechanisms and its practical application is not possible.
As the closest species to the OVF7 strain under investigation were Pseudomonas veronii and Pseudomonas fildensis, representatives of the group of fluorescent pseudomonads, we decided to analyze the degradative characteristics of strains of these species in the literature. Pavlov M.S., et al. first described a new species of P. fildesensis [58]. However, no literature sources describing hydrocarbon-degrading strains of the species Pseudomonas fildesensis have been found to date. In contrast, the species Pseudomonas veronii is known for its metabolic potential against hydrocarbons such as alkyl methyl ketones (Pseudomonas veronii strain MEK700 [59]); pentachlorophenol (Pseudomonas veronii strain PH-05 [60]); benzene and toluene (Pseudomonas veronii strain 1YdBTEX2 [61,62]); phenol (Pseudomonas veronii strain Ju-A1 [63]); 2,4,6-trinitrotoluene (Pseudomonas veronii strain S94 [64]); dibenzo-p-dioxin, dibenzofuran, and chlorodibenzo-p-dioxins (Pseudomonas veronii strain PH-03 [65]); 4-amylphenol and 4-hexylphenol (Pseudomonas strains INA04, INA05, and INA06 [66]); chlorobenzene (Pseudomonas veronii strain UFZ B549 [67]); 4-chorosalicylate (Pseudomonas veronii strain MT4 [68]); Pseudomonas veronii strain B547 [69], Pseudomonas veronii strain UFZ B549 [70,71], and Pseudomonas veronii strain 16-6A [72]); and naphthalene and n-decane (Pseudomonas veronii strain 7–41 [8]). It should be noted that the level of naphthalene degradation in strain 7–41 was comparable to that of the studied strain OVF7 (about 70%).
The number of studies on the degradative properties of microorganisms is increasing every year. Such studies are important for the development of bioremediation approaches. However, the degradation mechanism of oil hydrocarbons by both individual strains and by their consortia is still the subject of active studies. The uniqueness of the strain studied lies in its multi-degradation ability with respect to different classes of hydrocarbons. Therefore, at the stage of the physiological characterization of the strain, we tried to extrapolate a model experiment with a mixture of the different classes of hydrocarbons to the conditions under which microorganisms degrade such complex compounds as crude oil and oil products. There is no doubt that in such complex substances there are various physico-chemical interactions and the mutual influences between different classes of hydrocarbons on each other. Such chemical processes may also have a direct influence on the mechanism of pollutant degradation. The toxic effects of most hydrocarbons are due to a general, non-specific effect on membrane fluidity due to their accumulation in the lipid bilayer, causing an increase in membrane fluidity, resulting in non-specific membrane permeability. Most compounds with high hydrophobicity, such as alkanes, PAHs, and biphenyls, have very low solubility in water, so their bioavailability is too low to exhibit toxic effects. [73]. However, we observed a specific effect of bacterial cells sticking together as aggregates when cultured in an aqueous medium containing a hydrophobic carbon source. To rule out possible toxic effects of xenobiotics on cells and to determine cell viability, a live/dead cell detection method was applied using the LIVE/DEAD™ BacLight™ Bacterial Viability Kit. This fluorescence microscopy method assesses the viability of bacterial populations based on the integrity of the cell membrane. Cells with damaged membranes, considered dead or dying, stained red, whereas cells with intact membranes stained green. Since almost the entire cell population appeared to be alive under fluorescence microscopy regardless of the carbon source, and scanning microscopy clearly showed an organized structure, we suggest that the formation of trophic bonds is a feature of this strain and not the result of toxic effects of xenobiotics. Such bacterial structures and the reconstruction of their organization on a hydrophobic carbon source have been studied in detail by Dmitriev, V.V. [74].
In the present work, it was shown that Pseudomonas sp. strain OVF7 is able to grow on aliphatic substrates of the C8-C12 series and on naphthalene, and data on the degradation of naphthalene, n-dodecane, and their mixture (Figure 3) were obtained. We assume that the catabolic operons observed in the course of this work (described in Section 3.2) are responsible for the degradation of naphthalene and n-dodecane in the genome of Pseudomonas sp. strain OVF7. Our assumption is based on the high similarity of the structures of these operons (order and arrangement of genes, similarity of nucleotide sequences of regions containing nah, sal, and alk genes, and similarity of deduced amino acid sequences) with the operons for the degradation of n-alkanes and naphthalene, for which the functionality of degradation genes has been confirmed [8,14,51,52]. In addition, the differences in the lag phase shown on the bacterial growth curves (Figure 3) suggest different toxicity and bioavailability of different classes of hydrocarbons. It is important to note that the mixture of hydrocarbons contributes to the change in the growth curve, especially in the early stages. We think that this is due to the intermolecular interactions between n-dodecane and naphthalene [75,76]. Such a phenomenon is likely to influence the toxicity and bioavailability of the hydrocarbon mixture with respect to the strain studied. Summarizing the results of the physiology of the studied strain during the experiments, it can be assumed that Pseudomonas sp. strain OVF7 is capable of more active naphthalene degradation compared to n-dodecane, which correlates with the research data of the Pseudomonas veronii strain 7–41 of Mullaeva et al. [8]. However, in contrast to the above bacterium, no strain death was observed in the bi-substrate system. Perhaps such an effect is due to the ability of the strain to synthesize biopolymers and form a biofilm.
Biofilm formation is a sequential process controlled by cellular, surface, and environmental factors. To date, the mechanisms and components of biofilms formed by Staphylococcus epidermidis have been well studied. It has been found that most S. epidermidis strains are capable of forming so-called ica-dependent biofilms. The main component of ica-dependent biofilms is a polysaccharide-polymer β-1,6-N acetylglucosoamine called polysaccharide intercellular adhesin (PIA). The icaABCD operon is responsible for the synthesis of PIA. It consists of four genes: icaA (1238 bp), icaD (305 bp), icaC (869 bp), and icaB (1238 bp), and is regulated by the repressor gene icaR. The icaA gene encodes the membrane protein IcaA, which has N-acetylglucosamine transferase activity. IcaA, with the participation of the IcaD protein, synthesizes PIA from uridine diphosphate N-acetylglucosamine [77]. Another known PIA is PNAG (poly-β-1,6-N-acetyl-D-glucosamine). The pgaABCD operon is responsible for its synthesis in Escherichia coli, for example. Functionally and genetically related loci are found in other Gram-negative bacteria, including Klebsiella pneumoniae, Yersinia spp., Bordetella spp., Pseudomonas fluorescens, Actinobacillus pleuropneumoniae, Burkholderia cepacian, and Aggregatibacter actinomycetemcomitans. PNAG production by E. coli, A. pleuropneumoniae, Acinetobacter baumannii, Bordetella spp., A. actinomycetemcomitans, and Yersinia pestis has been confirmed biochemically and/or immunologically [78].
The strain Pseudomonas sp. strain OVF7 was able to form biofilms when grown in mineral medium with the following substrates as carbon and energy sources: naphthalene, succinate, n-dodecane, and a mixture of naphthalene and n-dodecane. The genome of the Pseudomonas sp. strain OVF7 strain, which we studied using the Bacterial and Viral Bioinformatics Resource Center (BV-BRC) service, revealed the absence of the icaABCD operon (or its homologues) and the presence of the pgaABCD locus. The locus pgaABCD has a high degree of similarity to the Burkholderia cepacia strain J2315 and B. cepacia strain ATCC25416 genetic locus [78], which encodes proteins involved in the biosynthesis and export of PNAG.
In [79], it was shown that P. fluorescens biofilm growth was affected by cultivation conditions (semi-static or dynamic) and changes in nutrient availability. The authors suggested that the observed differences in biofilm formation represent a specific response of the bacteria to nutrient availability and composition. Dynamic conditions with high nutrient levels resulted in reduced biofilm formation, described by heterogeneously distributed clusters of cells. Biofilms grown at lower nutrient levels and high flow rates were fully developed flat homogeneous biofilms. Biofilms growing in nutrient-rich media produced more exopolymers, resulting in a significant elastic response, as determined by high biofilm viscosity. More rigid biofilm properties, characterized by a higher modulus of elasticity, were observed in biofilms grown in low nutrient conditions. The observed biofilm stiffness may be related to the lower levels of exopolymeric substances formed. When grown on naphthalene, the Pseudomonas sp. strain OVF7 formed a perforated biofilm, probably due to the need for more oxygen in the PAH biodegradation process compared to alkanes.
The present work demonstrates the adaptation features of Pseudomonas sp. strain OVF7 with respect to hydrocarbons, i.e., the formation of biofilms of different architectures, the large number of IS elements, and the presence in the genome of degradative operons responsible for the degradation of different classes of hydrocarbons.
5. Conclusions
The Pseudomonas sp. strain OVF 7 was classified as a group of fluorescent pseudomonads on the basis of whole-genome sequencing, ANI and DDH, and rpoD nucleotide sequence analysis, and is probably a member of a new species. Such a hypothesis will be further verified by a detailed polyphasic taxonomy using phenotypic, chemotaxonomic, genetic, and phylogenetic methods. The Pseudomonas sp. OVF 7 has multi-degradation properties against hydrocarbons of different classes (aliphatic and aromatic compounds) and is able to form biofilms. The biofilm architecture is different when the bacterium is grown in a liquid mineral medium with various carbon substrates. The latter probably shows the adaptive functions of the studied strain’s biofilm, including the resistance to toxic hydrocarbons in the process of their biodegradation.
References
- Krauss, M.; Wilcke, W.; Martius, C.; Bandeira, A.G.; Garcia, M.V.; Amelung, W. Atmospheric versus biological sources of polycyclic aromatic hydrocarbons (PAHs) in a tropical rain forest environment. Environ. Pollut. 2005, 135, 143–154. [Google Scholar] [CrossRef]
- Patel, A.B.; Shaikh, S.; Jain, K.R.; Desai, C.; Madamwar, D. Polycyclic Aromatic Hydrocarbons: Sources, Toxicity, and Remediation Approaches. Front. Microbiol. 2020, 11, 562813. [Google Scholar] [CrossRef]
- Pandolfo, E.; Barra Caracciolo, A.; Rolando, L. Recent Advances in Bacterial Degradation of Hydrocarbons. Water 2023, 15, 375. [Google Scholar] [CrossRef]
- Rojo, F. Degradation of alkanes by bacteria. Environ. Microbiol. 2009, 11, 2477–2490. [Google Scholar] [CrossRef]
- Brzeszcz, J.; Kaszycki, P. Aerobic bacteria degrading both n-alkanes and aromatic hydrocarbons: An undervalued strategy for metabolic diversity and flexibility. Biodegradation 2018, 29, 359–407. [Google Scholar] [CrossRef]
- Ivanova, A.A.; Mullaeva, S.A.; Sazonova, O.I.; Petrikov, K.V.; Vetrova, A.A. Current research on simultaneous oxidation of aliphatic and aromatic hydrocarbons by bacteria of genus Pseudomonas. Folia Microbiol. 2022, 67, 591–604. [Google Scholar] [CrossRef]
- Whyte, L.G.; Bourbonniere, L.; Greer, C.W. Biodegradation of petroleum hydrocarbons by psychrotrophic Pseudomonas strains possessing both alkane (alk) and naphthalene (nah) catabolic pathways. Appl. Environ. Microbiol. 1997, 63, 3719–3723. [Google Scholar] [CrossRef]
- Mullaeva, S.A.; Delegan, Y.A.; Streletskii, R.A.; Sazonova, O.I.; Petrikov, K.V.; Ivanova, A.A.; Dyatlov, I.A.; Shemyakin, I.G.; Bogun, A.G.; Vetrova, A.A. Pseudomonas veronii strain 7-41 degrading medium-chain n-alkanes and polycyclic aromatic hydrocarbons. Sci. Rep. 2022, 12, 20527. [Google Scholar] [CrossRef]
- Lee, Y.; Lee, Y.; Jeon, C.O. Biodegradation of naphthalene, BTEX, and aliphatic hydrocarbons by Paraburkholderia aromaticivorans BN5 isolated from petroleum-contaminated soil. Sci. Rep. 2019, 9, 860. [Google Scholar] [CrossRef]
- Qi, J.; Wang, B.; Li, J.; Ning, H.; Wang, Y.; Kong, W.; Shen, L. Genetic determinants involved in the biodegradation of naphthalene and phenanthrene in Pseudomonas aeruginosa PAO1. Environ. Sci. Pollut. Res. Int. 2015, 22, 6743–6755. [Google Scholar] [CrossRef]
- Smits, T.H.; Witholt, B.; van Beilen, J.B. Functional characterization of genes involved in alkane oxidation by Pseudomonas aeruginosa. Antonie Van Leeuwenhoek 2003, 84, 193–200. [Google Scholar] [CrossRef] [PubMed]
- Nogales, J.; García, J.L.; Díaz, E. Degradation of Aromatic Compounds in Pseudomonas: A Systems Biology View. In Aerobic Utilization of Hydrocarbons, Oils and Lipids: Handbook of Hydrocarbon and Lipid Microbiology; Rojo, F., Ed.; Springer: Cham, Switzerland, 2017. [Google Scholar] [CrossRef]
- Jiménez, J.I.; Nogales, J.; García, J.L.; Díaz, E. A Genomic View of the Catabolism of Aromatic Compounds in Pseudomonas. In Handbook of Hydrocarbon and Lipid Microbiology; Timmis, K.N., Ed.; Springer: Berlin/Heidelberg, Germany, 2010. [Google Scholar] [CrossRef]
- Izmalkova, T.Y.; Sazonova, O.I.; Nagornih, M.O.; Sokolov, S.L.; Kosheleva, I.A.; Boronin, A.M. The organization of naphthalene degradation genes in Pseudomonas putida strain AK5. Res. Microbiol. 2013, 164, 244–253. [Google Scholar] [CrossRef] [PubMed]
- Abbasian, F.; Lockington, R.; Megharaj, M.; Naidu, R. A Review on the Genetics of Aliphatic and Aromatic Hydrocarbon Degradation. Appl. Biochem. Biotechnol. 2016, 178, 224–250. [Google Scholar] [CrossRef]
- Van Beilen, J.B.; Panke, S.; Lucchini, S.; Franchini, A.G.; Rothlisberger, M.; Witholt, B. Analysis of Pseudomonas putida alkane-degradation gene clusters and flanking insertion sequences: Evolution and regulation of the alk genes. Microbiology 2001, 147, 1621–1630. [Google Scholar] [CrossRef]
- Palleroni, N.J.; Pieper, D.H.; Moore, E.R.B. Microbiology of Hydrocarbon-Degrading Pseudomonas. In Handbook of Hydrocarbon and Lipid Microbiology; Timmis, K.N., Ed.; Springer: Berlin/Heidelberg, Germany, 2010. [Google Scholar] [CrossRef]
- Smits, T.H.; Balada, S.B.; Witholt, B.; van Beilen, J.B. Functional analysis of alkane hydroxylases from gram-negative and gram-positive bacteria. J. Bacteriol. 2002, 184, 1733–1742. [Google Scholar] [CrossRef]
- Panasia, G.; Philipp, B. LaoABCR, a novel system for oxidation of long-chain alcohols derived from SDS and alkane degradation in Pseudomonas aeruginosa. Appl. Environ. Microbiol. 2018, 84, e00626-18. [Google Scholar] [CrossRef]
- Salari, M.; Rahmanian, V.; Hashemi, S.A.; Chiang, W.-H.; Lai, C.W.; Mousavi, S.M.; Gholami, A. Bioremediation Treatment of Polyaromatic Hydrocarbons for Environmental Sustainability. Water 2022, 14, 3980. [Google Scholar] [CrossRef]
- Parrilli, E.; Tutino, M.L.; Marino, G. Biofilm as an adaptation strategy to extreme conditions. Rend. Fis. Acc. Lincei 2022, 33, 527–536. [Google Scholar] [CrossRef]
- Vandana; Das, S. Cell surface hydrophobicity and petroleum hydrocarbon degradation by biofilm-forming marine bacterium Pseudomonas furukawaii PPS-19 under different physicochemical stressors. J. Hazard. Mater. 2023, 457, 131795. [Google Scholar] [CrossRef]
- Ausubel, F.M.; Brent, R.; Kingston, R.E.; Moore, D.D.; Seidman, J.G.; Smith, J.A.; Struhl, K. Short Protocols in Molecular Biology, 4th ed.; John Wiley and Sons, Inc.: New York, NY, USA, 1990. [Google Scholar]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comp. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Kolmogorov, M.; Yuan, J.; Lin, Y.; Pevzner, P.A. Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 2019, 37, 540–546. [Google Scholar] [CrossRef] [PubMed]
- Hiltemann, S.; Rasche, H.; Gladman, S.; Hotz, H.R.; Lariviere, D.; Blankenberg, D.; Jagtap, P.D.; Wollmann, T.; Bretaudeau, A.; Goue, N.; et al. Galaxy Training: A powerful framework for teaching! PLoS Comput. Biol. 2023, 19, e1010752. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Goker, M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 2019, 10, 2182. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Carbasse, J.S.; Peinado-Olarte, R.L.; Goker, M. TYGS and LPSN: A database tandem for fast and reliable genome-based classification and nomenclature of prokaryotes. Nucleic Acids Res. 2022, 50, D801–D807. [Google Scholar] [CrossRef]
- Ondov, B.D.; Treangen, T.J.; Melsted, P.; Mallonee, A.B.; Bergman, N.H.; Koren, S.; Phillippy, A.M. Mash: Fast genome and metagenome distance estimation using MinHash. Genome Biol. 2016, 17, 132. [Google Scholar] [CrossRef]
- Lagesen, K.; Hallin, P.; Rodland, E.A.; Staerfeldt, H.H.; Rognes, T.; Ussery, D.W. RNAmmer: Consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007, 35, 3100–3108. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.P.; Goker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef] [PubMed]
- Lefort, V.; Desper, R.; Gascuel, O. FastME 2.0: A Comprehensive, Accurate, and Fast Distance-Based Phylogeny Inference Program. Mol. Biol. Evol. 2015, 32, 2798–2800. [Google Scholar] [CrossRef] [PubMed]
- Farris, J.S. Estimating phylogenetic trees from distance matrices. Am. Nat. 1972, 6, 645–667. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Hahnke, R.L.; Petersen, J.; Scheuner, C.; Michael, V.; Fiebig, A.; Rohde, C.; Rohde, M.; Fartmann, B.; Goodwin, L.A.; et al. Complete genome sequence of DSM 30083(T), the type strain (U5/41(T)) of Escherichia coli, and a proposal for delineating subspecies in microbial taxonomy. Stand. Genom. Sci. 2014, 9, 2. [Google Scholar] [CrossRef]
- Lee, I.; Ouk Kim, Y.; Park, S.C.; Chun, J. OrthoANI: An improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 2016, 66, 1100–1103. [Google Scholar] [CrossRef]
- Olson, R.D.; Assaf, R.; Brettin, T.; Conrad, N.; Cucinell, C.; Davis, J.J.; Dempsey, D.M.; Dickerman, A.; Dietrich, E.M.; Kenyon, R.W.; et al. Introducing the Bacterial and Viral Bioinformatics Resource Center (BV-BRC): A resource combining PATRIC, IRD and ViPR. Nucleic Acids Res. 2023, 51, D678–D689. [Google Scholar] [CrossRef]
- Grant, J.R.; Enns, E.; Marinier, E.; Mandal, A.; Herman, E.K.; Chen, C.Y.; Graham, M.; Van Domselaar, G.; Stothard, P. Proksee: In-depth characterization and visualization of bacterial genomes. Nucleic Acids Res. 2023, 51, W484–W492. [Google Scholar] [CrossRef]
- Petrikov, K.V.; Delegan, Y.A.; Surin, A.K.; Ponamoreva, O.N.; Puntus, I.F.; Filonov, A.E.; Boronin, A.M. Glycolipids of Pseudomonas and Rhodococcus oil-degrading bacteria used in bioremediation preparations: Formation and structure. Process Biochem. 2013, 48, 931–935. [Google Scholar] [CrossRef]
- Sambrook, J.R. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: Cold Spring Habor, NY, USA, 2001. [Google Scholar]
- Chun, J.; Oren, A.; Ventosa, A.; Christensen, H.; Arahal, D.R.; da Costa, M.S.; Rooney, A.P.; Yi, H.; Xu, X.W.; De Meyer, S.; et al. Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int. J. Syst. Evol. Microbiol. 2018, 68, 461–466. [Google Scholar] [CrossRef] [PubMed]
- Jain, C.; Rodriguez, R.L.; Phillippy, A.M.; Konstantinidis, K.T.; Aluru, S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 2018, 9, 5114. [Google Scholar] [CrossRef] [PubMed]
- Girard, L.; Lood, C.; Hofte, M.; Vandamme, P.; Rokni-Zadeh, H.; van Noort, V.; Lavigne, R.; De Mot, R. The Ever-Expanding Pseudomonas Genus: Description of 43 New Species and Partition of the Pseudomonas putida Group. Microorganisms 2021, 9, 1766. [Google Scholar] [CrossRef]
- Yen, K.M.; Gunsalus, I.C. Plasmid gene organization: Naphthalene/salicylate oxidation. Proc. Natl. Acad. Sci. USA 1982, 79, 874–878. [Google Scholar] [CrossRef] [PubMed]
- Eaton, R.W. Organization and evolution of naphthalene catabolic pathways: Sequence of the DNA encoding 2-hydroxychromene-2-carboxylate isomerase and trans-o-hydroxybenzylidenepyruvate hydratase-aldolase from the NAH7 plasmid. J. Bacteriol. 1994, 176, 7757–7762. [Google Scholar] [CrossRef]
- Dunn, N.W.; Gunsalus, I.C. Transmissible plasmid coding early enzymes of naphthalene oxidation in Pseudomonas putida. J. Bacteriol. 1973, 114, 974–979. [Google Scholar] [CrossRef]
- Li, W.; Shi, J.; Wang, X.; Han, Y.; Tong, W.; Ma, L.; Liu, B.; Cai, B. Complete nucleotide sequence and organization of the naphthalene catabolic plasmid pND6-1 from Pseudomonas sp. strain ND6. Gene 2004, 336, 231–240. [Google Scholar] [CrossRef]
- Dennis, J.J.; Zylstra, G.J. Complete sequence and genetic organization of pDTG1, the 83 kilobase naphthalene degradation plasmid from Pseudomonas putida strain NCIB 9816-4. J. Mol. Biol. 2004, 341, 753–768. [Google Scholar] [CrossRef]
- Pacwa-Plociniczak, M.; Plaza, G.A.; Poliwoda, A.; Piotrowska-Seget, Z. Characterization of hydrocarbon-degrading and biosurfactant-producing Pseudomonas sp. P-1 strain as a potential tool for bioremediation of petroleum-contaminated soil. Environ. Sci. Pollut. Res. Int. 2014, 21, 9385–9395. [Google Scholar] [CrossRef]
- Stackebrandt, E.; Frederiksen, W.; Garrity, G.M.; Grimont, P.A.D.; Kampfer, P.; Maiden, M.C.J.; Nesme, X.; Rossello-Mora, R.; Swings, J.; Truper, H.G.; et al. Report of the ad hoc committee for the re-evaluation of the species definition in bacteriology. Int. J. Syst. Evol. Microbiol. 2002, 52, 1043–1047. [Google Scholar] [CrossRef]
- Kim, M.; Oh, H.S.; Park, S.C.; Chun, J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int. J. Syst. Evol. Microbiol. 2014, 64, 346–351. [Google Scholar] [CrossRef]
- Auch, A.F.; von Jan, M.; Klenk, H.P.; Goker, M. Digital DNA-DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Stand. Genom. Sci. 2010, 2, 117–134. [Google Scholar] [CrossRef]
- Pavlov, M.S.; Lira, F.; Martinez, J.L.; Olivares-Pacheco, J.; Marshall, S.H. Pseudomonas fildesensis sp. nov. a psychrotolerant bacterium isolated from Antarctic soil of King George Island, South Shetland Islands. Int. J. Syst. Evol. Microbiol. 2020, 70, 3255–3263. [Google Scholar] [CrossRef] [PubMed]
- Onaca, C.; Kieninger, M.; Engesser, K.H.; Altenbuchner, J. Degradation of alkyl methyl ketones by Pseudomonas veronii MEK700. J. Bacteriol. 2007, 189, 3759–3767. [Google Scholar] [CrossRef]
- Nam, I.H.; Chang, Y.S.; Hong, H.B.; Lee, Y.E. A novel catabolic activity of Pseudomonas veronii in biotransformation of pentachlorophenol. Appl. Microbiol. Biotechnol. 2003, 62, 284–290. [Google Scholar] [CrossRef]
- De Lima-Morales, D.; Chaves-Moreno, D.; Wos-Oxley, M.L.; Jauregui, R.; Vilchez-Vargas, R.; Pieper, D.H. Degradation of benzene by Pseudomonas veronii 1YdBTEX2 and 1YB2 is catalyzed by enzymes encoded in distinct catabolism gene clusters. Appl. Environ. Microbiol. 2016, 82, 167–173. [Google Scholar] [CrossRef]
- Morales, M.; Sentchilo, V.; Bertelli, C.; Komljenovic, A.; Kryuchkova-Mostacci, N.; Bourdilloud, A.; Linke, B.; Goesmann, A.; Harshman, K.; Segers, F.; et al. The Genome of the Toluene-Degrading Pseudomonas veronii Strain 1YdBTEX2 and Its Differential Gene Expression in Contaminated Sand. PLoS ONE 2016, 11, e0165850. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Zhang, Y.; Liu, C.; Su, Z.; Zhao, R.; Zhou, J. Low-temperature phenol-degrading microbial agent: Construction and mechanism. Arch. Microbiol. 2023, 205, 193. [Google Scholar] [CrossRef] [PubMed]
- Avila-Arias, H.; Avellaneda, H.; Garzon, V.; Rodriguez, G.; Arbeli, Z.; Garcia-Bonilla, E.; Villegas-Plazas, M.; Roldan, F. Screening for biosurfactant production by 2,4,6-trinitrotoluene-transforming bacteria. J. Appl. Microbiol. 2017, 123, 401–413. [Google Scholar] [CrossRef]
- Hong, H.B.; Nam, I.H.; Murugesan, K.; Kim, Y.M.; Chang, Y.S. Biodegradation of dibenzo-p-dioxin, dibenzofuran, and chlorodibenzo-p-dioxins by Pseudomonas veronii PH-03. Biodegradation 2004, 15, 303–313. [Google Scholar] [CrossRef]
- Ajithkumar, B.; Ajithkumar, V.P.; Iriye, R. Degradation of 4-amylphenol and 4-hexylphenol by a new activated sludge isolate of Pseudomonas veronii and proposal for a new subspecies status. Res. Microbiol. 2003, 154, 17–23. [Google Scholar] [CrossRef] [PubMed]
- Nestler, H.; Kiesel, B.; Kaschabek, S.R.; Mau, M.; Schlomann, M.; Balcke, G.U. Biodegradation of chlorobenzene under hypoxic and mixed hypoxic-denitrifying conditions. Biodegradation 2007, 18, 755–767. [Google Scholar] [CrossRef] [PubMed]
- Pawelczyk, S.; Abraham, W.R.; Harms, H.; Muller, S. Community-based degradation of 4-chorosalicylate tracked on the single cell level. J. Microbiol. Methods 2008, 75, 117–126. [Google Scholar] [CrossRef] [PubMed]
- Vogt, C.; Simon, D.; Alfreider, A.; Babel, W. Microbial degradation of chlorobenzene under oxygen-limited conditions leads to accumulation of 3-chlorocatechol. Environ. Toxicol. Chem. 2004, 23, 265–270. [Google Scholar] [CrossRef]
- Balcke, G.U.; Wegener, S.; Kiesel, B.; Benndorf, D.; Schlomann, M.; Vogt, C. Kinetics of chlorobenzene biodegradation under reduced oxygen levels. Biodegradation 2008, 19, 507–518. [Google Scholar] [CrossRef]
- Kiesel, B.; Balcke, G.U.; Dietrich, J.; Vogt, C.; Geyer, R. Microbial community shifts as a response to efficient degradation of chlorobenzene under hypoxic conditions. Biodegradation 2008, 19, 435–446. [Google Scholar] [CrossRef]
- Gobel, M.; Kranz, O.H.; Kaschabek, S.R.; Schmidt, E.; Pieper, D.H.; Reineke, W. Microorganisms degrading chlorobenzene via a meta-cleavage pathway harbor highly similar chlorocatechol 2,3-dioxygenase-encoding gene clusters. Arch. Microbiol. 2004, 182, 147–156. [Google Scholar] [CrossRef]
- Heipieper, H.J.; Martínez, P.M. Toxicity of hydrocarbons to microorganisms. In Handbook of Hydrocarbon and Lipid Microbiology; Timmis, K.N., Ed.; Springer: Berlin/Heidelberg, Germany, 2010; pp. 1563–1573. ISBN 978-3-540-77587-4. [Google Scholar]
- Dmitriev, V.V.; Crowley, D.; Rogachevsky, V.V.; Negri, C.M.; Rusakova, T.G.; Kolesnikova, S.A.; Akhmetov, L.I. Microorganisms form exocellular structures, trophosomes, to facilitate biodegradation of oil in aqueous media. FEMS Microbiol. Lett. 2011, 315, 134–140. [Google Scholar] [CrossRef]
- Leshchev, S.M.; Sin’kevich, A.V. Comparative assessment of the solvating powers of solvents of different nature with respect to condensed aromatic hydrocarbons. Russ. J. Appl. Chem. 2003, 76, 1483–1488. [Google Scholar] [CrossRef]
- Yakubouski, S.; Bulauka, Y.; Kazak, Y. Comparative evaluation of the solvating power of hydrocarbons and alcohols towards naphthalene. Russ. J. Bull. Polotsk State Univ. Ser. B Ind. Appl. Sci. Chem. Technol. Labour Prot. 2016, 3, 160–163. [Google Scholar]
- Gerke, C.; Kraft, A.; Sussmuth, R.; Schweitzer, O.; Gotz, F. Characterization of the N-acetylglucosaminyltransferase activity involved in the biosynthesis of the Staphylococcus epidermidis polysaccharide intercellular adhesin. J. Biol. Chem. 1998, 273, 18586–18593. [Google Scholar] [CrossRef] [PubMed]
- Yakandawala, N.; Gawande, P.V.; LoVetri, K.; Cardona, S.T.; Romeo, T.; Nitz, M.; Madhyastha, S. Characterization of the poly-beta-1,6-N-acetylglucosamine polysaccharide component of Burkholderia biofilms. Appl. Environ. Microbiol. 2011, 77, 8303–8309. [Google Scholar] [CrossRef] [PubMed]
- Allen, A.; Habimana, O.; Casey, E. The effects of extrinsic factors on the structural and mechanical properties of Pseudomonas fluorescens biofilms: A combined study of nutrient concentrations and shear conditions. Colloids Surf. B Biointerfaces 2018, 165, 127–134. [Google Scholar] [CrossRef] [PubMed]

