1. Introduction
Lichens are the only organisms on Earth that consist of fungi (the mycobiont) and photosynthetic cyanobacteria or algae (the photobiont) in a permanent symbiotic relationship. The symbiotic relationships between fungi and algae have been found to exist for at least 400 million years [1]. The fungal partner is considered when determining the identity of lichens, and as of now, ascomycetes have been identified as the fungal partner in the majority of lichen species [2,3]. Thus, “lichen-forming fungi (LFF)” refer to fungi that establish a symbiotic relationship and live in a lichen thallus during the entire life cycle.
Lichens can be found in almost all places on Earth, including the harshest environments, such as extreme cold and hot, covering over 10% of the terrestrial surface [4,5,6,7,8]. Lichens’ broad habitats and long lifespans demand a robust defense against harmful environmental elements and predatory species. One line of defense is the biosynthesis of secondary metabolites, chemicals produced by an organism that are not required for its growth, development, and reproduction but have biological activity that can indirectly equip the organism to defend better and survive. It is well known that lichens produce a wide range of secondary metabolites, and harnessing their potential for human benefit has received much attention [9,10,11].
Attempts are often made to find the genes involved in secondary metabolite synthesis [12]. Among the genes identified as key players in the synthesis of metabolites, tailoring enzymes such as cytochrome P450 monooxygenases (CYPs/P450s) were found to be critical in not only synthesis but also in conferring diversity to the metabolites [12]. P450s are heme-thiolate-containing proteins present in almost all organisms, including non-living entities such as viruses [13,14]. P450s are well known in biology due to their stereo- and regio-specific oxidation of diverse compounds, including their synthesis and contributing diversity to the secondary metabolites in organisms [15,16].
For P450s, a distinct nomenclature and classification scheme has been devised [17,18]. The prefix “CYP” stands for cytochrome P450 monooxygenase, and it is followed by an Arabic numeral indicating the family, a capital letter indicating the subfamily, and an Arabic digit representing the individual P450 in a family. According to the annotation/classification criterion, all P450s with >40% identity belong to the same family, and all P450s with >55% identity belong to the same subfamily.
P450s in fungi are involved in primary and secondary metabolism, where, due to their key enzymatic reactions, they serve frequently as a drug target [19,20,21]. Concerning secondary metabolism, fungal P450s are known to be involved in synthesizing many secondary metabolites with potential biotechnological applications [19,22,23]. It is now well-established that P450s play a crucial role in an organism’s adaptation vis à vis the lifestyle of organisms that impact the P450 content in their genome [24]. A recent study published by our laboratory reported this phenomenon in Pezizomycetes [25]. Despite belonging to the same class, saprophytes and ectomycorrhizal species were found to have diverse P450 complements characteristic of their lifestyles [25].
A handful of studies on lichens reported the involvement of P450s in synthesizing secondary metabolites, where P450s were found to carry out a critical enzymatic reaction [26,27,28,29]. Despite the importance of P450s in the biosynthesis of secondary metabolites in lichens, to date, a systematic analysis of P450s in these organisms has yet to be reported. Thus, this study is aimed to address this research gap.
In this study, we have selected five lichens from the fungal class Lecanoromycetes . General information on these five Lecanoromycetes is presented in . The criteria for selecting these five Lecanoromycetes is that their genomes were well annotated, published, and available for public use at the Joint Genome Institute’s MycoCosm site [30]. Furthermore, detailed information on the secondary metabolite biosynthetic gene clusters (BGCs) and the genes forming the BGCs is also available. This allows us to easily identify P450s, carry out their annotation and phylogenetic analysis, and identify their role in secondary metabolism.
As part of this study, we also named characterized lichen P450s reported in the literature to enable researchers to use the proper P450 names. Last, we also compared P450s between Lecanoromycetes and Pezizomycetes to see any commonalities or diversity as they belong to the same phylum, Ascomycota [36].
2. Materials and Methods
2.1. Species and Databases
The study employed five Lecanoromycetes . All Lecanoromycetes genomes used in the study have been published and are freely accessible to the public via the Joint Genome Institute’s MycoCosm site [30].
2.2. Genome Data Mining and Identification of P450s
Genome data mining and identification of P450s in Lecanoromycetes was carried out following the method described elsewhere [25]. Each set of annotated proteins from Lecanoromycetes was briefly searched for P450s using the InterPro code “IPR001128”. The hit protein sequences were downloaded and screened for P450 motifs, such as EXXR and CXG [37,38].
Proteins containing one of these motifs or short in length (fewer than 350 amino acids) were classified as P450 fragments. In comparison, proteins containing fewer than 350 amino acids but showing a similarity to P450s were classified as partial P450s. Proteins with all of the P450 characteristic motifs were considered P450s. P450 family and subfamily analyses were performed on proteins grouped as P450s. The P450 fragments that were <350 amino acids long were considered incomplete. These sequences were produced by annotation pipelines that are in use by the genome producers. We rely on that software for the accurate assembly of the protein sequences. When the sequences are short, there is a high probability that they come from pseudogenes as the same software accurately predicted full-length P450 proteins from the same genome. Furthermore, the DNA sequence was manually copied and pasted such that 5000 bp upstream and downstream were taken, and genes were predicted using GENSCAN [39].
2.3. Assigning P450 Family and Subfamily
We used a BLAST (Basic Local Alignment Search Tool) analysis of Lecanoromycetes P450s against all fungal sequences listed on the Cytochrome P450 Homepage [18] to estimate the percentage identity with named homolog P450s in order to determine P450 families and subfamilies. The proteins were then classified into distinct P450 families and subfamilies following the International P450 Nomenclature standards [17,18]. Proteins with >40% and >55% amino acid identity were classified as belonging to the same P450 family and subfamily. New P450 families were assigned to proteins that had less than 40% similarity with the closest P450 homologs. The P450s are included in , together with the names given to them and the P450 fragment sequences.
2.4. Phylogenetic Analysis
Phylogenetic analysis of P450s was carried out following the method described elsewhere [25]. The P450 protein sequences were aligned using the MAFFT v6.864 [40] program with an automatically optimized model option available at the T-REX web server [41]. The alignments were then automatically subjected to inference and optimization of the tree by the Trex web server [41] with its embedded weighting procedure. Finally, the best-inferred tree was visualized, colored, and generated by the Interactive Tree Of Life (iTOL) [42].
2.5. P450 Family Conservation Analysis
Heat maps were created from the P450 family data using the method described in the literature [25]. The data were shown as −3 for the absence of a gene (green) and +3 for the presence of a gene (red). A tab-delimited file was imported into the multi-experiment viewer (MeV) [43]. The data were clustered using a Euclidean distance metric and hierarchical clustering. The vertical axis was made up of Lecanoromycetes, and the horizontal axis was made up of P450 families.
2.6. Identification of P450s That Are Part of Secondary Metabolite Biosynthetic Gene Clusters
P450s that are part of secondary metabolite biosynthetic gene clusters (BGCs), were identified following the method described elsewhere [25] with slight modification. Each of the BGCs listed in the Joint Genome Institute’s MycoCosm site [30] for each of the species was manually searched for gene/protein sequences in that cluster. If a P450 was listed as a part of the cluster, the P450 protein ID and its sequence were noted and matched with its assigned name. The cluster ID, cluster type, scaffold information (genomic location), and P450s part of the cluster were presented in table format as a standard practice.
2.7. Assigning the P450 Family and Subfamilies to the Functionally Characterized Lichen P450s
A handful of P450s from lichens are functionally characterized and shown to be involved in synthesizing different secondary metabolites [26,27,28,29]. The P450 sequences from these lichens were retrieved from the literature, and P450 families and P450 subfamilies were assigned to these P450s, as indicated in Section 2.3.
3. Results and Discussion
3.1. Lecanoromycetes Have More P450s than Pezizomycetes
A genome-wide analysis of P450s in the five Lecanoromycetes yielded 584 hit proteins (Figure 1A). Further examination of hit proteins for typical P450 motifs (as indicated in Section 2.2) revealed that not all hit proteins are P450s. Among the hits, 434 have all the typical P450 motifs and are considered P450s; of the rest, 124 hit proteins were identified as P450 fragments and 26 hit proteins as partial P450s (Figure 1A). A species-level analysis indicated that Cladonia grayi Cgr/DA2myc/ss and Umbilicaria pustulata have no partial P450s (Figure 1B).
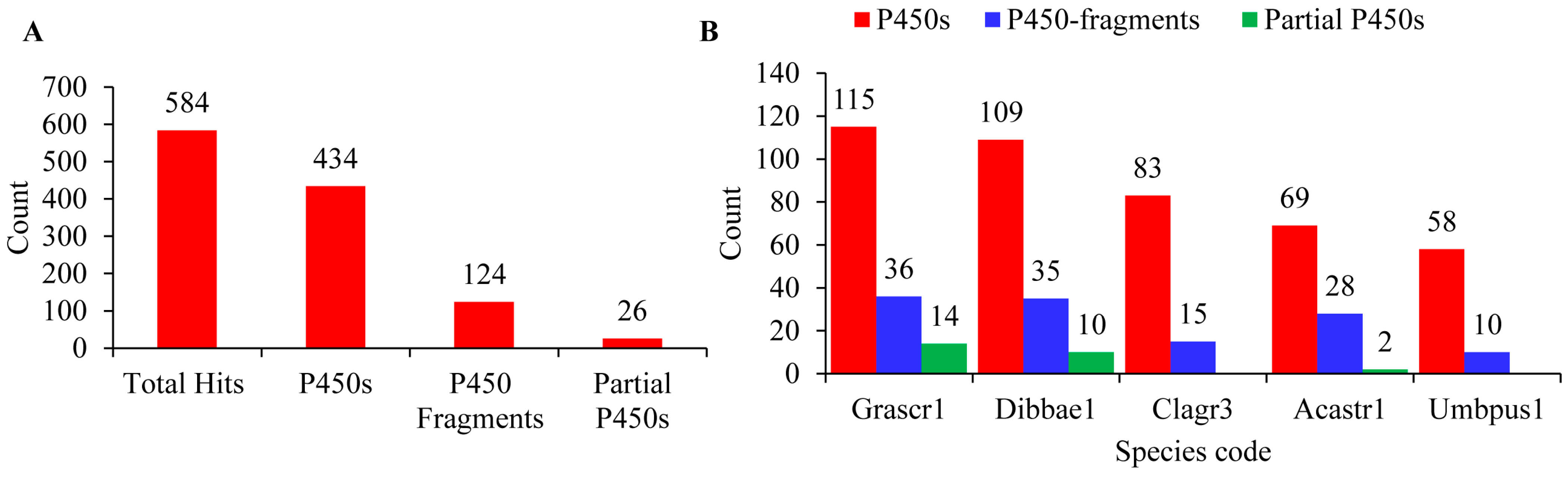
Figure 1. Analysis of P450s in the five Lecanoromycetes. Identification of P450s (A) and species level comparison of P450s (B) in the five Lecanoromycetes. Species codes: Grascr1, Graphis scripta CBS 132367; Dibbae1, Dibaeis baeomyces; Clagr3, Cladonia grayi Cgr/DA2myc/ss; Acastr1, Acarospora Strigata CBS 132363; Umbpus1, Umbilicaria pustulata.
The number of P450s in the five Lecanoromycetes ranged from 58 to 115 P450s, with an average of 87 P450s. Among Lecanoromycetes, Graphis scripta CBS 132367 has the highest number of P450s (115), and U. pustulata has the lowest number of P450s (58) in their genome (Figure 1B). The average number of P450s in Lecanoromycetes is 2.5 times higher than the Pezizomycetes [25], suggesting that Lecanoromycetes have more P450s in their genome. A list of P450s identified in Lecanoromycetes, along with their assigned name and protein IDs, are presented in .
3.2. Lecanoromycetes Have Higher P450 Family Diversity than Pezizomycetes
The 434 P450s identified in the five Lecanoromycetes were categorized into 178 P450 families and 345 P450 subfamilies following the International P450 Nomenclature Committee’s criteria [17] and the phylogenetic analysis (Figure 2 and . As stated in Section 2.3, P450s were allocated to various P450 families and subfamilies based on the percent sequence identity; however, phylogenetic analysis is crucial in determining which subfamilies belong to P450s that are borderline with the identified homolog P450s and fall into the range of about 55% identical. These borderline P450s were assigned to the correct subfamilies based on alignment on the evolutionary tree. Additionally, phylogenetic analysis can be used to determine evolutionary links, such as how closely two species’ P450 genes are related.
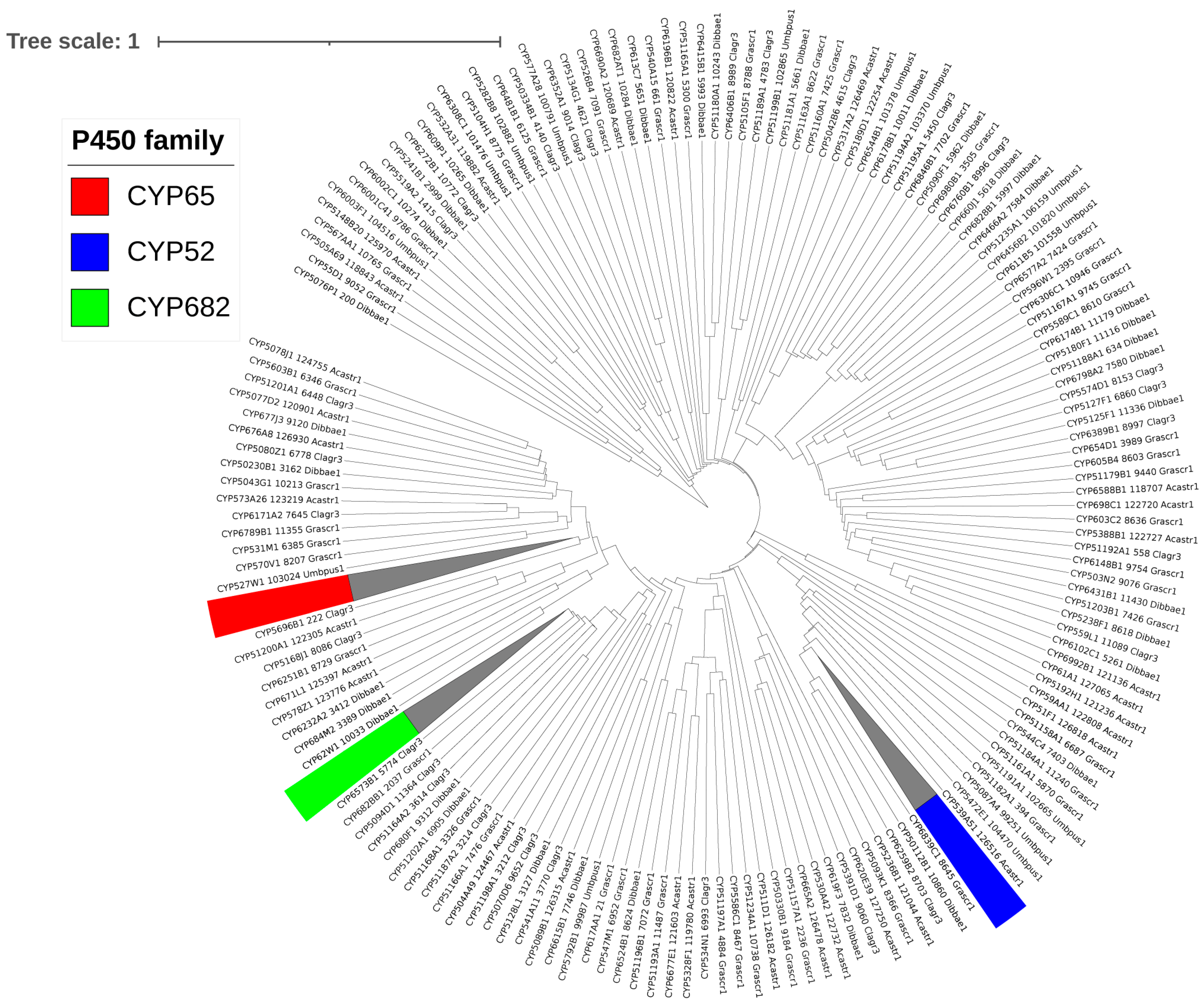
Figure 2. Phylogenetic analysis of Lecanoromycetes P450s. A single P450 from each of the 178 P450 families was used to construct the tree, except for P450 families with ten or more members highlighted in different colors; all members were included, and the branch collapsed. A high-quality figure with all 434 P450s is presented in .
Only 6 P450 families, CYP65, CYP59, CYP52, CYP682, CYP584, and CYP6001, have ten or more members among the 178 identified families in the five Lecanoromycetes. As a result, it is safe to conclude that P450 families in Lecanoromycetes have not bloomed (a single P450 family with many genes) . It is worth noting that the same phenomenon was found in Pezizomycetes, where no P450 family bloomed [25]. In contrast to what was found for Lecanoromycetes and Pezizomycetes, P450 family blooming was observed in several fungal species [21,44]. P450 subfamily-level blooming was also not observed for Lecanoromycetes, whereas the blooming of two P450 subfamilies was observed for Pezizomycetes [25]. Compared to the 19 Pezizomycetes, the 5 Lecanoromycetes have more P450 families (153 vs. 178), indicating higher P450 family diversity in the Lecanoromycetes.
The number of P450 families ranged from 45 to 84, with an average of 63 P450 families, and the number of P450 subfamilies ranged from 112 to 57, with an average of 84 P450 families (Figure 3 and ). G. scripta CBS 132367 and U. pustulata had the highest and lowest number of P450 families and P450 subfamilies in their genomes (Figure 3 and ). Lecanoromycetes have a higher number of P450 families and P450 subfamilies per species compared to Pezizomycetes [25], indicating that species belonging to the former group have the highest P450 family and subfamily diversity.
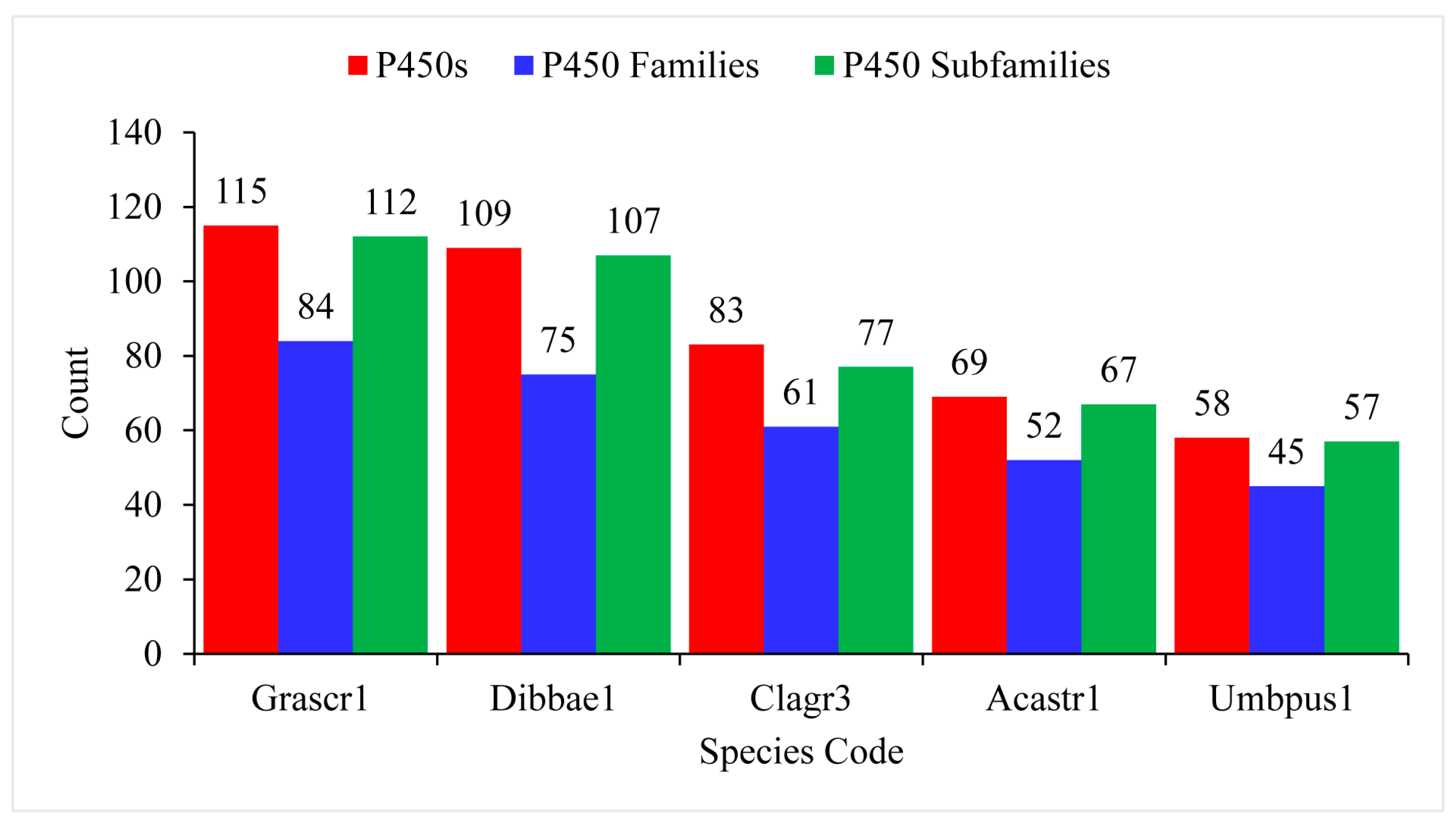
Figure 3. Comparative analysis of P450s, P450 families, and P450 subfamilies in five the Lecanoromycetes. Detailed information is presented in . Species codes: Grascr1, Graphis scripta CBS 132367; Dibbae1, Dibaeis baeomyces; Clagr3, Cladonia grayi Cgr/DA2myc/ss; Acastr1, Acarospora Strigata CBS 132363; Umbpus1, Umbilicaria pustulata.
A comparative analysis of P450s profiles between Lecanoromycetes and Pezizomycetes revealed that species belonging to these classes have different P450 profiles with few similarities (Figure 4). Despite both classes belonging to the same phylum, Ascomycota [36], and subphylum, Pezizomycotina, only 34 P450 families are common, and quite a large number of P450 families were found to be unique to Lecanoromycetes (144 P450 families) and Pezizomycetes (119 P450 families) (Figure 4). This indicates that after speciation, the lifestyle or ecological niches played a key role in shaping the P450 content in these species, as observed for bacterial [24,45] and some fungal species [21,44,46].
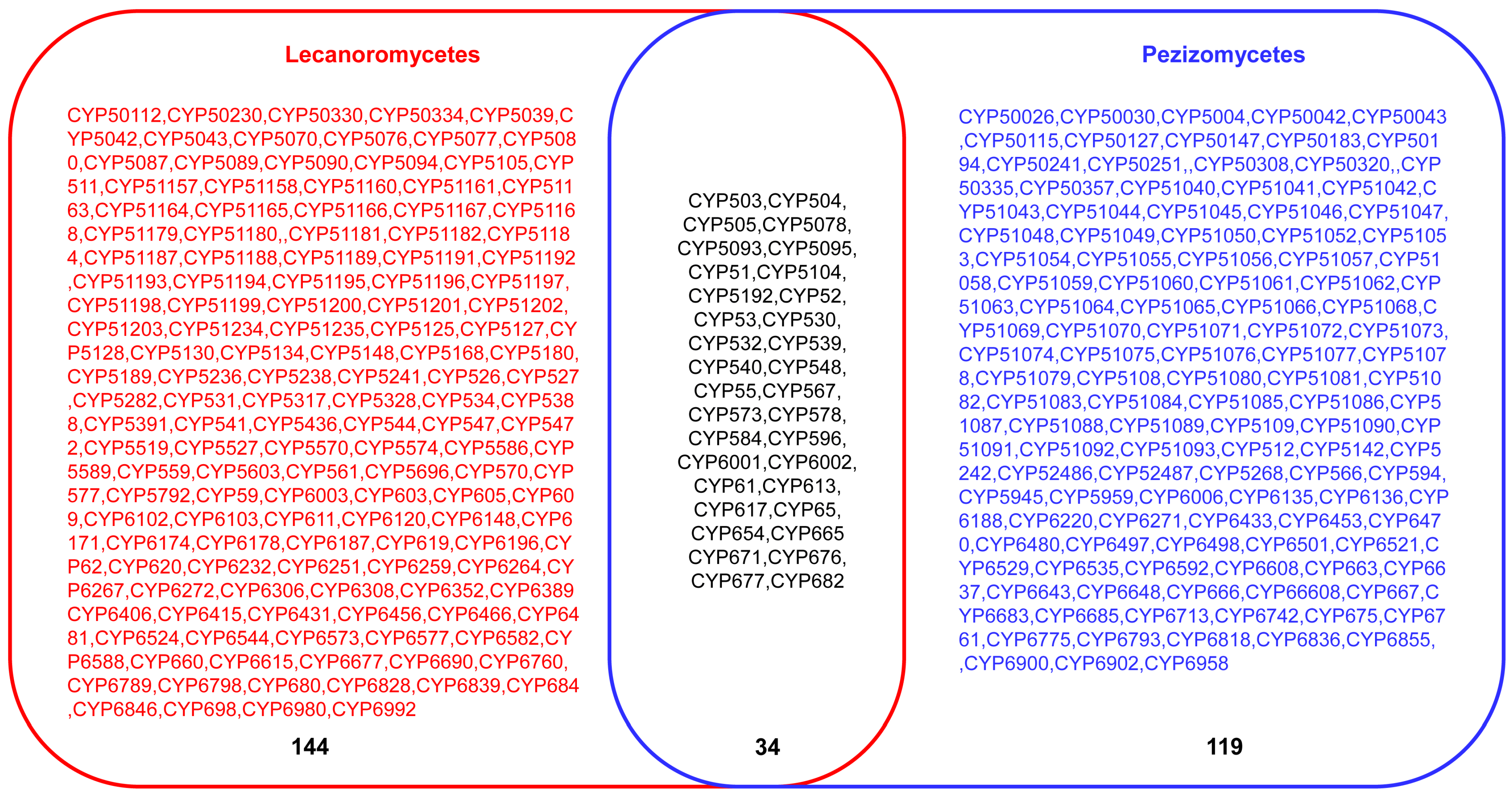
Figure 4. Comparative analysis of P450 families between Lecanoromycetes and Pezizomycetes. The number indicates the total number of P450 families.
3.3. Fifteen P450 Families Are Conserved in the Five Lecanoromycetes
P450 family conservation analysis revealed that only 15 out of 178 families, namely, CYP51, CYP52, CYP532, CYP534, CYP53, CYP544, CYP548, CYP584, CYP59, CYP6001, CYP617, CYP61, CYP65, CYP6677, and CYP682, are conserved in five Lecanoromycetes (Figure 5). The conservation of these P450 families across the five Lecanoromycetes suggests that these P450 families may play important roles. It is well established that CYP51 and CYP61 are involved in sterol biosynthesis [47,48,49], which is required for cell wall membranes, and that CYP6001 members are involved in fatty acid oxidation [50]. CYP53 members play a key role in detoxifying toxic molecules in fungi; thus, CYP53 has been proposed as a common alternative anti-fungal drug target [21]. CYP52 members are involved in the oxidation of alkanes and fatty acids and the biosynthesis of sophorolipids [51,52]. CYP65 members are involved in the biosynthesis of sesquiterpenoid mycotoxins [53]. From the characterized homolog P450s functions, it is clear that members belonging to these P450 families play an important role in fungi and, thus, one can expect the conservation of these P450 family members in Lecanoromycetes.
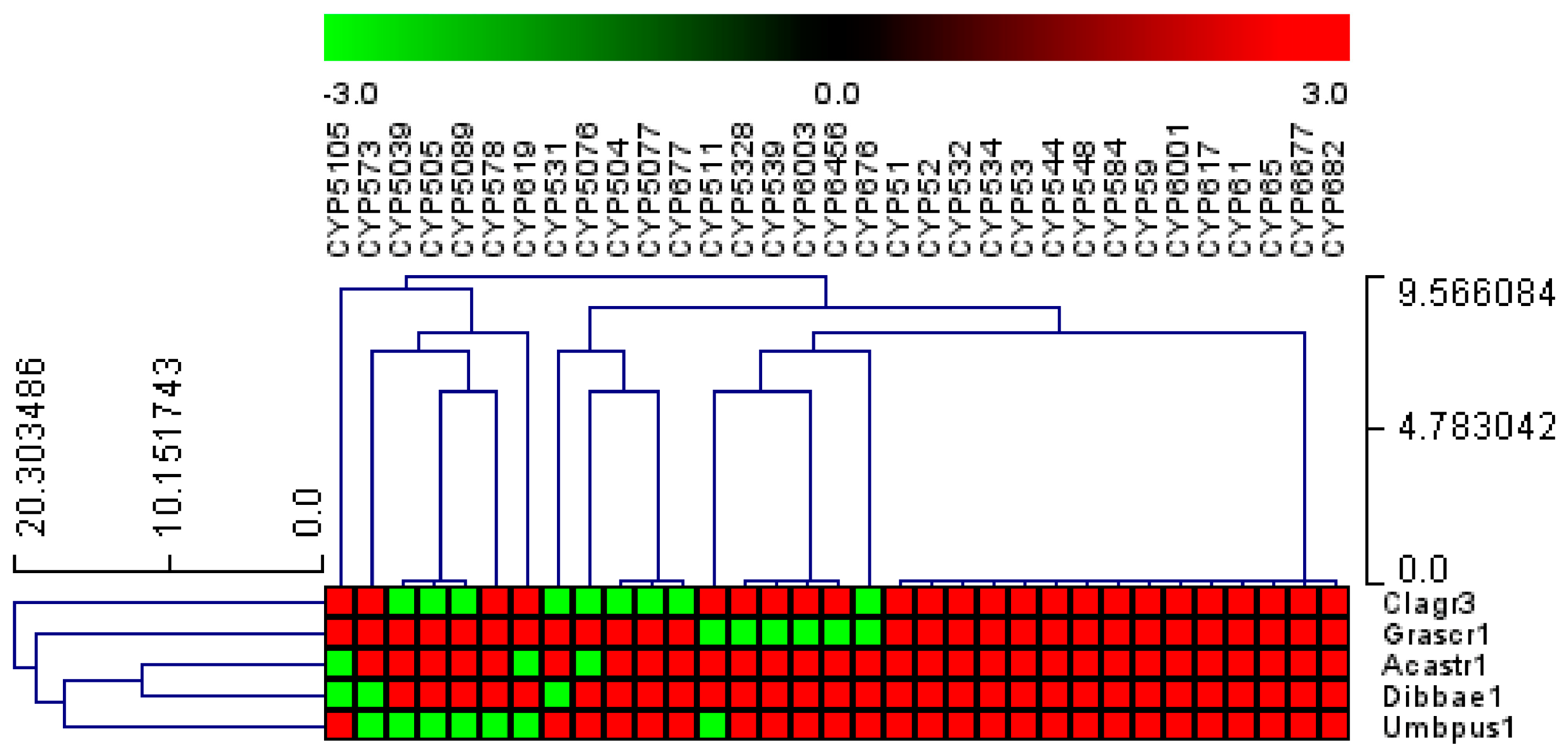
Figure 5. Analysis of P450 family conservation in the five Lecanoromycetes. P450 families that are conserved in three or more species are presented. The heat map shows that the P450 family is either present (red) or absent (green) in Lecanoromycetes. The horizontal axis is made up of P450 families, and the vertical axis is made up of Lecanoromycetes. provides a thorough examination of P450 family conservation in Lecanoromycetes.
3.4. Fifty-Five P450s from the Five Lecanoromycetes Are Involved in the Biosynthesis of Secondary Metabolites
A biosynthetic gene cluster (BGC) analysis revealed that 73 P450s are part of different BGCs, indicating their possible role in synthesizing secondary metabolites . Among the 73 P450s, 18 were P450-fragments/partial P450s, and thus only 55 P450s can be assumed to be involved in the biosynthesis of secondary metabolites . Among the five Lecanoromycetes, G. scripta CBS 132367 have the highest number of P450s that are part of BGCs (19 P450s), followed by C. grayi Cgr/DA2myc/ss v2.0 (13 P450s), Dibaeis baeomyces (12 P450s), A. strigata CBS 132363 (9 P450s), and U. pustulata (2 P450s).
The analysis of P450 families that are part of BGCs revealed that the CYP65 family has the highest number of members (seven members) followed by CYP5039 (five members) and CYP5077 (three members). Eight P450 families have only two members, and twenty-four P450 families have only a single member as part of BGCs .
As reported in the literature, the functional characterization of a handful of P450s from lichens revealed that P450s perform a critical catalytic reaction in synthesizing secondary metabolites [26,27,28,29] (Figure 6). These P450 sequences were retrieved from the literature and annotated in this study to enable their correct identification. These P450s and their assigned names and protein sequences are presented in .
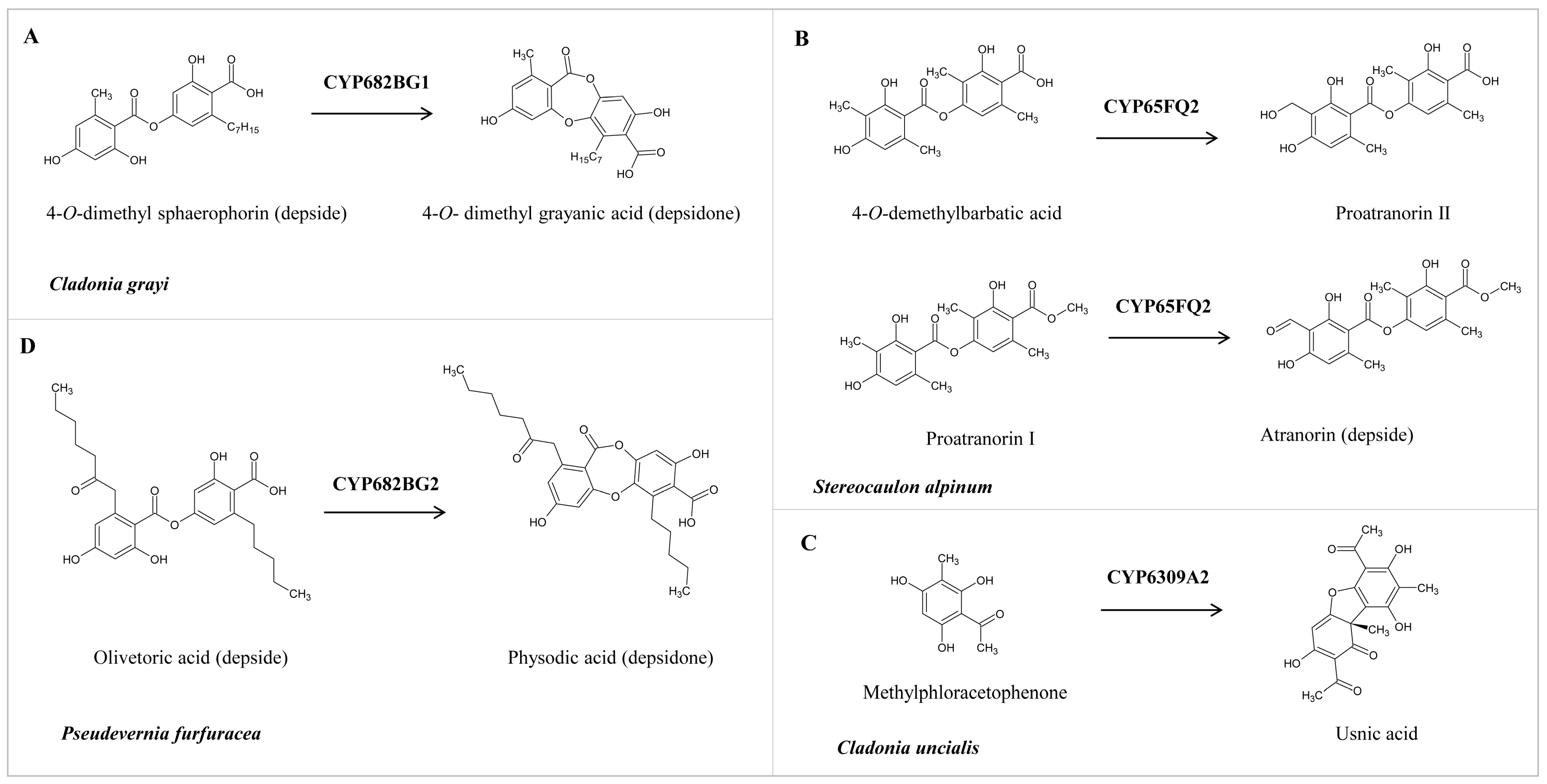
Figure 6. Role of lichen P450s in the biosynthesis of different secondary metabolites. (A,B) Conversion of depside to depsidone by CYP682BG1 and CYP682BG2. (C) Oxidation of 4-O-demethylbarbatic acid and proatranorin I by CYP65Q2. (D) Oxidative dimerization of methylphloracetophenone to usnic acid by CYP6309A2.
To enable researchers to identify the P450 and its functions, we provide detailed information on lichen P450s and their elucidated functions. CYP682BCG1 from C. grayi performs the oxidative coupling of 4-O-demethyl sphaerophorin (depside) to 4-O-demethyl grayanic acid (depsidone) via ether bond formation (Figure 6A) [26]. CYP65FQ2 from Stereocaulon alpinum catalyzes the oxidation of C-9 of 4-O-demethylbarbatic acid and proatranorin I to generate proatranorin II and atranorin (ß-orcinol depside), respectively (Figure 6B) [27]. CYP6309A2, named methylphloracetophenone oxidase, from Cladonia uncialis, performs the oxidative dimerization of methylphloracetophenone to usnic acid (dibezofurane derivative) (Figure 6C) [28]. CYP682BG1 from Pseudevernia furfuracea (physodic acid chemotype) performs the oxidative coupling of olivetoric acid (depside), leading to physodic acid (depsidone) synthesis via ether bond formation (Figure 6D) [29]. It is interesting to note that CYP682BG in C. grayi and P. furfuracea converts depside to depsidone irrespective of the side chains in depside.
4. Conclusions
The quest for novel compounds with potential biotechnological value is increasing, and researchers are probing previously unexplored venues. Due to their adaptability to practically all climatic conditions, lichens generate a broad spectrum of natural compounds that may help them protect themselves against infections or predators. Several studies have indicated that these compounds have potential biotechnological values. A few have been shown to act as anti-bacterial, anti-fungal, and anti-cancer agents. A handful of studies have tried to outline the biosynthetic pathway of these compounds and shed light on genes involved in synthesizing these compounds. Among these genes, tailoring enzymes such as P450s have been found to play a key role in synthesizing these compounds in lichens. The results from this study provide important information on these enzymes in lichens. Also, this study indicates that speciation and adaptation to diverse ecological niches led to the development of a diverse P450 complement in Lecanoromycetes and Pezizomycetes. More lichen genomes are under investigation for P450 profiles to understand evolutionary aspects and assess their role in secondary metabolism. Considering the unique reactions performed by lichen P450s, one can predict that a great potential for lichen P450s is waiting to be discovered.
References
- Lücking, R.; Nelsen, M.; Krings, M.; Harper, C.; Cúneo, N.; Rothwell, G. Transformative Paleobotany; Papers to Commemorate the Life and Legacy of Thomas N. Taylor; Elsevier: Amsterdam, The Netherlands, 2018; pp. 551–590. [Google Scholar]
- Grube, M.; Wedin, M. Lichenized fungi and the evolution of symbiotic organization. Microbiol. Spectr. 2016, 4, 34. [Google Scholar] [CrossRef] [PubMed]
- Sipman, H.J.; Aptroot, A. Where are the missing lichens? Mycol. Res. 2001, 105, 1433–1439. [Google Scholar] [CrossRef]
- Ellis, C.J. Lichen epiphyte diversity: A species, community and trait-based review. Perspect. Plant Ecol. Evol. Syst. 2012, 14, 131–152. [Google Scholar] [CrossRef]
- Lee, Y.M.; Kim, E.H.; Lee, H.K.; Hong, S.G. Biodiversity and physiological characteristics of Antarctic and Arctic lichens-associated bacteria. World J. Microbiol. Biotechnol. 2014, 30, 2711–2721. [Google Scholar] [CrossRef]
- Kranner, I.; Beckett, R.; Hochman, A.; Nash III, T.H. Desiccation-tolerance in lichens: A review. Bryologist 2008, 111, 576–593. [Google Scholar] [CrossRef]
- Nguyen, K.-H.; Chollet-Krugler, M.; Gouault, N.; Tomasi, S. UV-protectant metabolites from lichens and their symbiotic partners. Nat. Prod. Rep. 2013, 30, 1490–1508. [Google Scholar] [CrossRef] [PubMed]
- Boustie, J.; Tomasi, S.; Grube, M. Bioactive lichen metabolites: Alpine habitats as an untapped source. Phytochem. Rev. 2011, 10, 287–307. [Google Scholar] [CrossRef]
- Ren, M.; Jiang, S.; Wang, Y.; Pan, X.; Pan, F.; Wei, X. Discovery and excavation of lichen bioactive natural products. Front. Microbiol. 2023, 14, 1177123. [Google Scholar] [CrossRef]
- Varol, M. Lichens as a promising source of unique and functional small molecules for human health and well-being. Stud. Nat. Prod. Chem. 2019, 60, 425–458. [Google Scholar]
- Gautam, A.K.; Yadav, D.; Bhagyawant, S.S.; Singh, P.K.; Jin, J.-O. Lichen: A comprehensive review on Lichens as a natural sources exploring nutritional and biopharmaceutical benefits. Prog. Nutr. 2021, 23. [Google Scholar] [CrossRef]
- Singh, G. Linking lichen metabolites to genes: Emerging concepts and lessons from molecular biology and metagenomics. J. Fungi 2023, 9, 160. [Google Scholar] [CrossRef] [PubMed]
- Ngcobo, P.E.; Nkosi, B.V.Z.; Chen, W.; Nelson, D.R.; Syed, K. Evolution of cytochrome P450 enzymes and their redox partners in Archaea. Int. J. Mol. Sci. 2023, 24, 4161. [Google Scholar] [CrossRef] [PubMed]
- Lamb, D.C.; Follmer, A.H.; Goldstone, J.V.; Nelson, D.R.; Warrilow, A.G.; Price, C.L.; True, M.Y.; Kelly, S.L.; Poulos, T.L.; Stegeman, J.J. On the occurrence of cytochrome P450 in viruses. Proc. Natl. Acad. Sci. USA 2019, 116, 12343–12352. [Google Scholar] [CrossRef] [PubMed]
- Podust, L.M.; Sherman, D.H. Diversity of P450 enzymes in the biosynthesis of natural products. Nat. Prod. Rep. 2012, 29, 1251–1266. [Google Scholar] [CrossRef]
- Greule, A.; Stok, J.E.; De Voss, J.J.; Cryle, M.J. Unrivalled diversity: The many roles and reactions of bacterial cytochromes P450 in secondary metabolism. Nat. Prod. Rep. 2018, 35, 757–791. [Google Scholar] [CrossRef]
- Nelson, D.R. Cytochrome P450 nomenclature, 2004. Methods Mol. Biol. 2006, 320, 1–10. [Google Scholar] [CrossRef]
- Nelson, D.R. The cytochrome p450 homepage. Hum. Genom. 2009, 4, 59–65. [Google Scholar] [CrossRef]
- Črešnar, B.; Petrič, Š. Cytochrome P450 enzymes in the fungal kingdom. Biochim. Biophys. Acta BBA-Proteins Proteom. 2011, 1814, 29–35. [Google Scholar] [CrossRef]
- Lepesheva, G.I.; Friggeri, L.; Waterman, M.R. CYP51 as drug targets for fungi and protozoan parasites: Past, present and future. Parasitology 2018, 145, 1820–1836. [Google Scholar] [CrossRef]
- Jawallapersand, P.; Mashele, S.S.; Kovacic, L.; Stojan, J.; Komel, R.; Pakala, S.B.; Krasevec, N.; Syed, K. Cytochrome P450 monooxygenase CYP53 family in fungi: Comparative structural and evolutionary analysis and its role as a common alternative anti-fungal drug target. PLoS ONE 2014, 9, e107209. [Google Scholar] [CrossRef]
- Keller, N.P. Fungal secondary metabolism: Regulation, function and drug discovery. Nat. Rev. Microbiol. 2019, 17, 167–180. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Guo, J.; Cheng, F.; Li, S. Cytochrome P450 enzymes in fungal natural product biosynthesis. Nat. Prod. Rep. 2021, 38, 1072–1099. [Google Scholar] [CrossRef] [PubMed]
- Msweli, S.; Chonco, A.; Msweli, L.; Syed, P.R.; Karpoormath, R.; Chen, W.; Gront, D.; Nkosi, B.V.Z.; Nelson, D.R.; Syed, K. Lifestyles shape the cytochrome P450 repertoire of the Bacterial phylum Proteobacteria. Int. J. Mol. Sci. 2022, 23, 5821. [Google Scholar] [CrossRef]
- Nsele, N.N.; Padayachee, T.; Nelson, D.R.; Syed, K. Pezizomycetes genomes reveal diverse P450 complements characteristic of saprotrophic and ectomycorrhizal lifestyles. J. Fungi 2023, 9, 830. [Google Scholar] [CrossRef] [PubMed]
- Armaleo, D.; Sun, X.; Culberson, C. Insights from the first putative biosynthetic gene cluster for a lichen depside and depsidone. Mycologia 2011, 103, 741–754. [Google Scholar] [CrossRef]
- Kim, W.; Liu, R.; Woo, S.; Kang, K.B.; Park, H.; Yu, Y.H.; Ha, H.-H.; Oh, S.-Y.; Yang, J.H.; Kim, H. Linking a gene cluster to atranorin, a major cortical substance of lichens, through genetic dereplication and heterologous expression. mBio 2021, 12, e0111121. [Google Scholar] [CrossRef]
- Abdel-Hameed, M.; Bertrand, R.L.; Piercey-Normore, M.D.; Sorensen, J.L. Putative identification of the usnic acid biosynthetic gene cluster by de novo whole-genome sequencing of a lichen-forming fungus. Fungal Biol. 2016, 120, 306–316. [Google Scholar] [CrossRef]
- Singh, G.; Armaleo, D.; Dal Grande, F.; Schmitt, I. Depside and depsidone synthesis in lichenized fungi comes into focus through a genome-wide comparison of the olivetoric acid and physodic acid chemotypes of Pseudevernia furfuracea. Biomolecules 2021, 11, 1445. [Google Scholar] [CrossRef]
- Kuo, A.; Salamov, A.; Korzeniewski, F.; Nordberg, H.; Shabalov, I.; Dubchak, I.; Otillar, R.; Riley, R.; Ohm, R.; Nikitin, R.; et al. MycoCosm portal: Gearing up for 1000 fungal genomes. Nucleic Acids Res. 2013, 42, D699–D704. [Google Scholar] [CrossRef]
- McDonald, T.R.; Mueller, O.; Dietrich, F.S.; Lutzoni, F. High-throughput genome sequencing of lichenizing fungi to assess gene loss in the ammonium transporter/ammonia permease gene family. BMC Genom. 2013, 14, 225. [Google Scholar] [CrossRef]
- Armaleo, D.; Müller, O.; Lutzoni, F.; Andrésson, Ó.S.; Blanc, G.; Bode, H.B.; Collart, F.R.; Dal Grande, F.; Dietrich, F.; Grigoriev, I.V. The lichen symbiosis re-viewed through the genomes of Cladonia grayi and its algal partner Asterochloris glomerata. BMC Genom. 2019, 20, 605. [Google Scholar] [CrossRef] [PubMed]
- Tanahashi, T.; Kuroishi, M.; Kuwahara, A.; Nagakura, N.; Hamada, N. For phenolics from the cultured lichen mycobiont of Graphis scripta var. pulverulenta. Chem. Pharm. Bull. 1997, 45, 1183–1185. [Google Scholar] [CrossRef]
- Greshake Tzovaras, B.; Segers, F.H.; Bicker, A.; Dal Grande, F.; Otte, J.; Anvar, S.Y.; Hankeln, T.; Schmitt, I.; Ebersberger, I. What is in Umbilicaria pustulata? A metagenomic approach to reconstruct the holo-genome of a lichen. Genome Biol. Evol. 2020, 12, 309–324. [Google Scholar] [CrossRef]
- Dal Grande, F.; Sharma, R.; Meiser, A.; Rolshausen, G.; Büdel, B.; Mishra, B.; Thines, M.; Otte, J.; Pfenninger, M.; Schmitt, I. Adaptive differentiation coincides with local bioclimatic conditions along an elevational cline in populations of a lichen-forming fungus. BMC Evol. Biol. 2017, 17, 93. [Google Scholar] [CrossRef] [PubMed]
- Schoch, C.L.; Sung, G.-H.; López-Giráldez, F.; Townsend, J.P.; Miadlikowska, J.; Hofstetter, V.; Robbertse, B.; Matheny, P.B.; Kauff, F.; Wang, Z. The Ascomycota tree of life: A phylum-wide phylogeny clarifies the origin and evolution of fundamental reproductive and ecological traits. Syst. Biol. 2009, 58, 224–239. [Google Scholar] [CrossRef] [PubMed]
- Gotoh, O. Substrate recognition sites in cytochrome P450 family 2 (CYP2) proteins inferred from comparative analyses of amino acid and coding nucleotide sequences. J. Biol. Chem. 1992, 267, 83–90. [Google Scholar] [CrossRef]
- Syed, K.; Mashele, S.S. Comparative analysis of P450 signature motifs EXXR and CXG in the large and diverse kingdom of fungi: Identification of evolutionarily conserved amino acid patterns characteristic of P450 family. PLoS ONE 2014, 9, e95616. [Google Scholar] [CrossRef]
- Burge, C.B.; Karlin, S. Finding the genes in genomic DNA. Curr. Opin. Struct. Biol. 1998, 8, 346–354. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Kuma, K.; Toh, H.; Miyata, T. MAFFT version 5: Improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005, 33, 511–518. [Google Scholar] [CrossRef]
- Boc, A.; Diallo, A.B.; Makarenkov, V. T-REX: A web server for inferring, validating and visualizing phylogenetic trees and networks. Nucleic Acids Res. 2012, 40, W573–W579. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016, 44, W242–W245. [Google Scholar] [CrossRef]
- Saeed, A.I.; Sharov, V.; White, J.; Li, J.; Liang, W.; Bhagabati, N.; Braisted, J.; Klapa, M.; Currier, T.; Thiagarajan, M.; et al. TM4: A free, open-source system for microarray data management and analysis. BioTechniques 2003, 34, 374–378. [Google Scholar] [CrossRef] [PubMed]
- Ngwenya, M.L.; Chen, W.; Basson, A.K.; Shandu, J.S.; Yu, J.H.; Nelson, D.R.; Syed, K. Blooming of unusual cytochrome P450s by tandem duplication in the pathogenic fungus Conidiobolus coronatus. Int. J. Mol. Sci. 2018, 19, 1711. [Google Scholar] [CrossRef] [PubMed]
- Zondo, N.M.; Padayachee, T.; Nelson, D.R.; Syed, K. Saprophytic to pathogenic mycobacteria: Loss of cytochrome P450s vis a vis their prominent involvement in natural metabolite biosynthesis. Int. J. Mol. Sci. 2022, 24, 149. [Google Scholar] [CrossRef]
- Kgosiemang, I.K.R.; Syed, K.; Mashele, S.S. Comparative genomics and evolutionary analysis of cytochrome P450 monooxygenases in fungal subphylum Saccharomycotina. J. Pure Appl. Microbiol. 2014, 8, 12. [Google Scholar]
- Lamb, D.C.; Hargrove, T.Y.; Zhao, B.; Wawrzak, Z.; Goldstone, J.V.; Nes, W.D.; Kelly, S.L.; Waterman, M.R.; Stegeman, J.J.; Lepesheva, G.I. Concerning P450 evolution: Structural analyses support bacterial origin of sterol 14α-demethylases. Mol. Biol. Evol. 2021, 38, 952–967. [Google Scholar] [CrossRef] [PubMed]
- Kelly, S.L.; Lamb, D.C.; Baldwin, B.C.; Corran, A.J.; Kelly, D.E. Characterization of Saccharomyces cerevisiae CYP61, sterol delta22-desaturase, and inhibition by azole antifungal agents. J. Biol. Chem. 1997, 272, 9986–9988. [Google Scholar] [CrossRef]
- Chen, W.; Lee, M.-K.; Jefcoate, C.; Kim, S.-C.; Chen, F.; Yu, J.-H. Fungal cytochrome p450 monooxygenases: Their distribution, structure, functions, family expansion, and evolutionary origin. Genome Biol. Evol. 2014, 6, 1620–1634. [Google Scholar] [CrossRef]
- Brodhun, F.; Gobel, C.; Hornung, E.; Feussner, I. Identification of PpoA from Aspergillus nidulans as a Fusion Protein of a Fatty Acid Heme Dioxygenase/Peroxidase and a Cytochrome P450. J. Biol. Chem. 2009, 284, 11792–11805. [Google Scholar] [CrossRef]
- Ortiz-Álvarez, J.; Becerra-Bracho, A.; Méndez-Tenorio, A.; Murcia-Garzón, J.; Villa-Tanaca, L.; Hernández-Rodríguez, C. Phylogeny, evolution, and potential ecological relationship of cytochrome CYP52 enzymes in Saccharomycetales yeasts. Sci. Rep. 2020, 10, 10269. [Google Scholar] [CrossRef]
- Huang, F.-C.; Peter, A.; Schwab, W. Expression and characterization of CYP52 genes involved in the biosynthesis of sophorolipid and alkane metabolism from Starmerella bombicola. Appl. Environ. Microbiol. 2014, 80, 766–776. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M.; Tokai, T.; Takahashi-Ando, N.; Ohsato, S.; Fujimura, M. Molecular and genetic studies of Fusarium trichothecene biosynthesis: Pathways, genes, and evolution. Biosci. Biotechnol. Biochem. 2007, 71, 2105–2123. [Google Scholar] [CrossRef] [PubMed]

