1. Introduction
Staphylococcus aureus (S. aureus) is a Gram-positive coccus that causes significant infections worldwide, including bacteremia, endocarditis, osteomyelitis, and skin and soft tissue infections, due to its easy transmission and commensal nature [1]. Not only S. aureus but also coagulase-negative Staphylococci (CoNS), which currently are defined as more than 40 species, are frequently associated with opportunistic human infections. S. epidermidis and S. haemolyticus are the major species of CoNS frequently isolated from clinical specimens [2]. Furthermore, Mammaliicoccus sciuri (previously called S. sciuri) is part of the normal flora of goats and camels, and it is a rare opportunistic pathogen in humans [3]. S. aureus possesses a unique set of virulence factors, including toxins, enzymes, and metallophores, which enable it to survive extreme conditions, promote tissue colonization, cause systemic infection, and evade the host’s immunity [4]. By utilizing metallophores, this bacterium can sequester metal ions from its environment [5]. S. aureus infections have previously been treated with beta-lactams, including penicillin and, later, methicillin, as well as sulfonamides, tetracyclines, and macrolides [6]. However, antibiotic-resistant strains of S. aureus have developed due to repeated exposure to antibiotics, leading to an increase in methicillin-resistant S. aureus (MRSA) infections globally. MRSA is one of the most causative pathogens of surgical site infections (SSIs), and it is a prevalent bacterium that frequently colonizes hospital environments and causes hospital-acquired infections [6] and community-acquired infections [7].
MRSA is characterized by resistance to penicillins, cephalosporins, and carbapenems, with the exception of the new anti-MRSA cephalosporins ceftaroline and ceftobiprole antimicrobial agents [6,8]. The main mechanism of resistance is an altered penicillin-binding protein (PBP2a/c) encoded by the mecA gene [1]. The mecA gene is regarded as the gold standard for identifying isolates of MRSA, and it is a helpful marker. It is highly conserved in staphylococcal strains and acquired through horizontal gene transfer. mecA is carried/located on the mobile genetic element staphylococcal cassette chromosome (SCC) mec, and it codes for the low-affinity PBP2a [9]. Other chromosomal factors, such as the high-level expression of femA and femB, also seem to be essential for high-level methicillin resistance [10]. The current treatment options for more serious MRSA infections requiring hospitalization include parenteral antimicrobials, such as teicoplanin, tigecycline, linezolid [8], trimethoprim–sulfamethoxazole, doxycycline, daptomycin [6], and vancomycin [8]. However, the majority of MRSA strains are capable of evolution and have acquired resistance to a variety of antibiotics, including those listed above [9,11].
Vancomycin resistance in MRSA was first discovered in 1996 in Japan following a few years of commercializing the antibiotic [12]. Vancomycin resistance is acquired through mutations and cell wall modification [12,13] mediated by a vanA gene cluster that can be acquired from vancomycin-resistant enterococcus (VRE) [11] through mobile genetic elements like transposonTn1546. Vancomycin-resistant S. aureus (VRSA) infections are treated with antibiotics like tigecycline, quinupristin, daptomycin, ceftobiprole, iclaprim, linezolid, and new glycopeptides (telavancin, oritavancin, and dabavancin) [14].
Globally, the prevalence of VRSA was 16% in Africa, 1% in Europe, 3% in South America, 4% in North America, and 5% in Asia [15]. A systematic review and meta-analysis revealed a highly variable prevalence of VRSA and MRSA in Ethiopian S. aureus isolates. The MRSA prevalence ranged from 8.3% to 77.3% (with a pooled prevalence of 32.5%) [16]. In the same way, there was a 5.1% to 44.3% variation in VRSA prevalence. [17]. These days, MRSA is considered a serious threat to public health, and it is one of the pathogens that needs to be treated with high priority. However, the molecular epidemiology of MRSA and VRSA is less well documented in Ethiopia, and published reports on MRSA- and VRSA-causing SSIs are scarce. Furthermore, almost all earlier reports depend on phenotypic laboratory methods. Therefore, in this study, we used the Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry (MALDI-TOF MS) technique for the confirmation of bacterial isolates and multiplex polymerase chain reaction (PCR) for the detection of mecA, femA, vanA, and vanB.
2. Results
In this study, a total of 752 participants were included. S. aureus was isolated from 21.7% (163) of these participants. Following that, a cefoxitin disc diffusion test was used as a substitute marker for oxacillin and other penicillinase-resistant penicillins to ascertain the percentage of MRSA.
Of all participants, 5.3% (40/752) carried bacteria characterized as MRSA, while among isolates of S. aureus, the frequency of MRSA was 24.5% . All methicillin-resistant isolates were also tested for vancomycin susceptibility. Except for one isolate (2.5%), all tested isolates for vancomycin were sensitive.
The age of study participants with MRSA ranged from 8 days to 85 years, with a mean age (±standard deviation) of 35 ± 28.3 years and a median of 30 years, and 58.3% (95) were males. Fifty-nine (36.2%) of the participants received antimicrobial prophylaxis before the procedure, and 47.2% (63) underwent surgeries lasting longer than an hour .
The likelihood of MRSA SSI occurrence was about 3.7 times higher among patients aged ≥ 61 (AOR = 3.729 (1.179–11.791)) compared to those aged ≤ 60. Similarly, the relative risk of MRSA SSI occurrence was about 1.9 times more likely among patients who had a hospital stay ≥ 7 days (AOR = 1.856 (0.688–5.311)). Also, those who had a history of antibiotic use had a 3.7 times higher risk of developing methicillin-resistant Staphylococci infections (AOR = 3.692 (1.059–2.800)) than methicillin-sensitive S. aureus (MSSA) SSI. The likelihood of SSI occurrence was about 3.16 times more likely among patients who had antimicrobial prophylaxis during the operation (AOR = 3.066 (1.101–9.392)) than those who had antimicrobial prophylaxis before the operation. All p-value < 0.05 .
2.1. MALDI-TOF MS Identification of Methicillin-Resistant Staphylococcus Isolates
Of the 40 phenotypic MRSA bacterial isolates, MALDI-TOF MS only identified 77.5% (31/40) as S. aureus, while 6 were identified as M. sciuri, and the other three as S. warneri, S. epidermidis, and S. haemolyticus (Figure 1). The majority (70%) of methicillin-resistant Staphylococcus isolates were identified from Debre Tabor Comprehensive Specialized Hospital (Figure 2) with 47.5% as S. aureus, 15% as M. sciuri, 2.5% as S. warneri, 2.5% as S. epidermidis and 2.5% as S. haemolyticus.
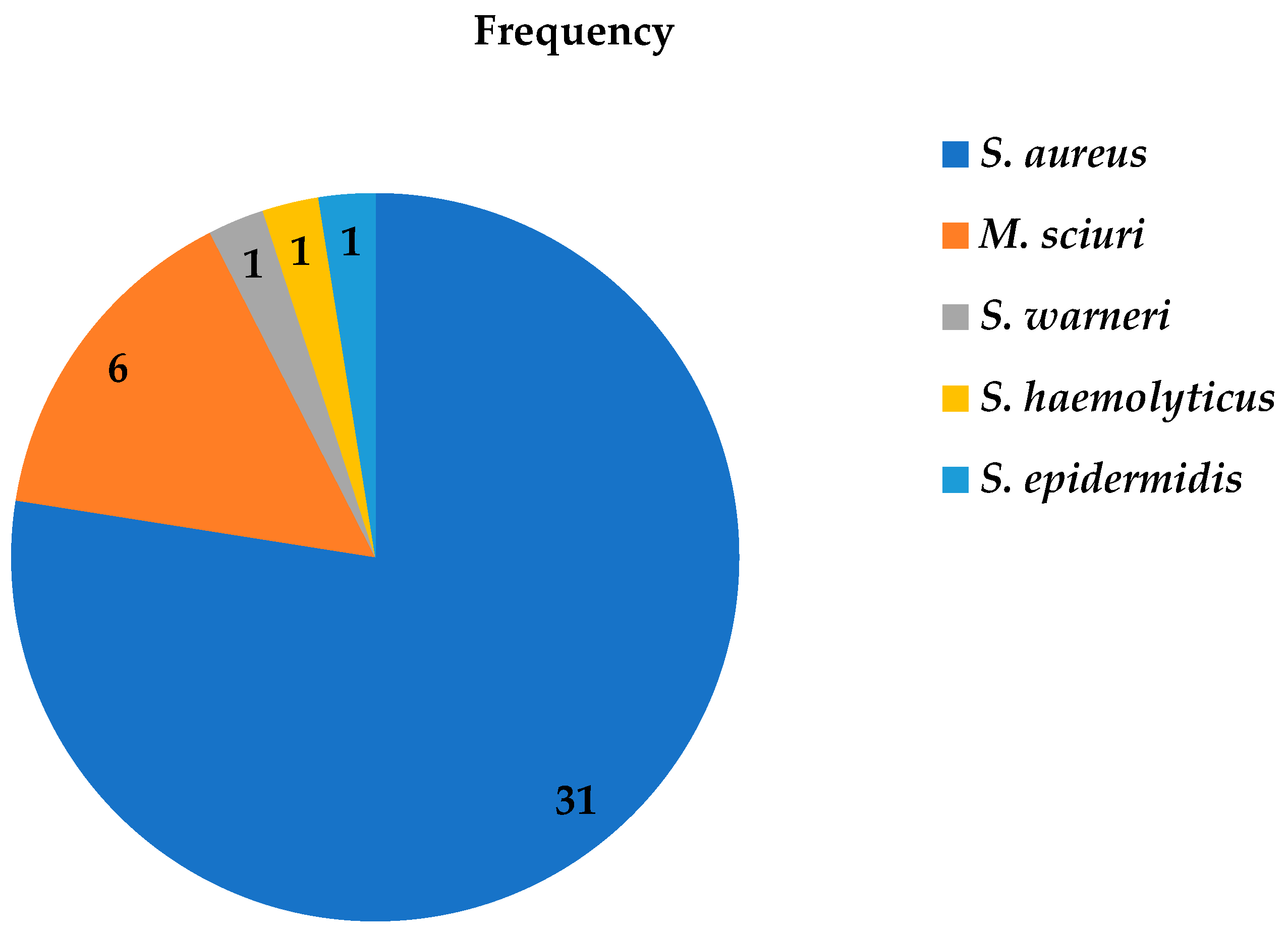
Figure 1. Frequency of methicillin-resistant Staphylococci isolates from patients diagnosed with surgical site infection at four different hospitals in Ethiopia using MALDI-TOF MS between July 2020 and August 2021.
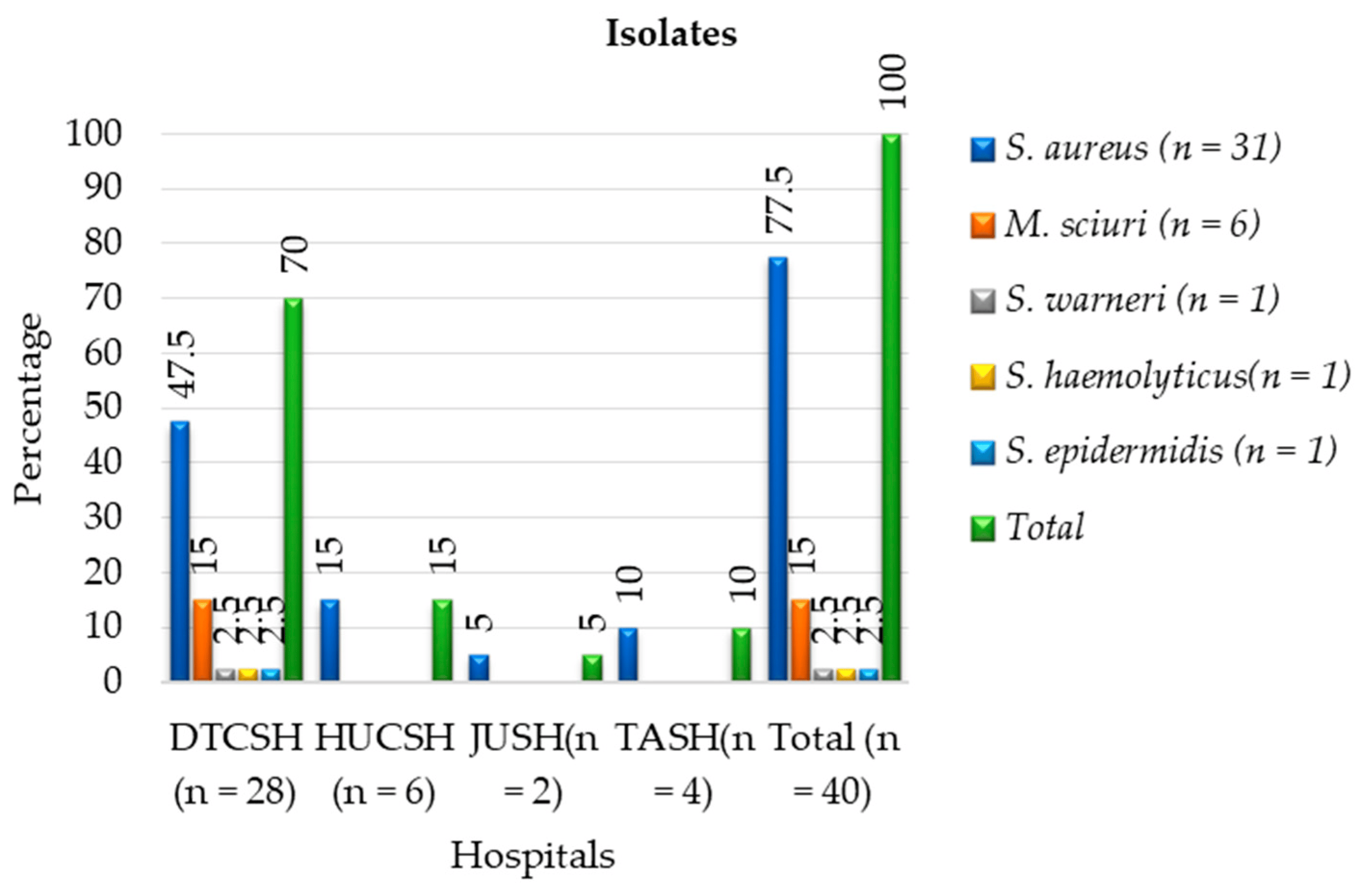
Figure 2. Frequencies of MALDI-TOF MS identification and distribution of phenotypic methicillin-resistant Staphylococci and M. sciuri isolates at each hospital between July 2020 and August 2021. DTCSH: Debre Tabor Comprehensive Specialized Hospital; HUCSH: Hawassa University Comprehensive Specialized Hospital; JUTSH: Jimma University Teaching Specialized Hospital; TASH: Tikur Anbessa Specialized Hospital.
2.2. PCR amplification of mecA, femA, van A, and vanB
Detection of mecA, femA, van A, and vanB was performed for all MRSA and methicillin-resistant Staphylococci other than S. aureus (MRSOSA). The PCR tests revealed that 27.5% (11/40) contained the mecA gene, 25% (10/40) were both mecA- and femA- positive, and 92.5% (37/40) showed the femA gene (Figure 3). On the other hand, from all eleven isolates that contained the mecA gene only, four were S. aureus, whereas five were M. sciuri, one was S. warneri, and one was S. haemolyticus, respectively (Figure 4 and . Among S. aureus isolates, only 12.9% (4/31) carried the mecA gene (MRSA), whereas 83.3% (5/6) of M. sciuri and both S. warneri and S. haemolyticus isolates carried mecA (Figure 3).

Figure 3. Frequency and distribution of cefoxitin resistance and PCR-confirmed gene among each Staphylococci isolate in patient diagnosed with surgical site infection between July 2020 and August 2021.
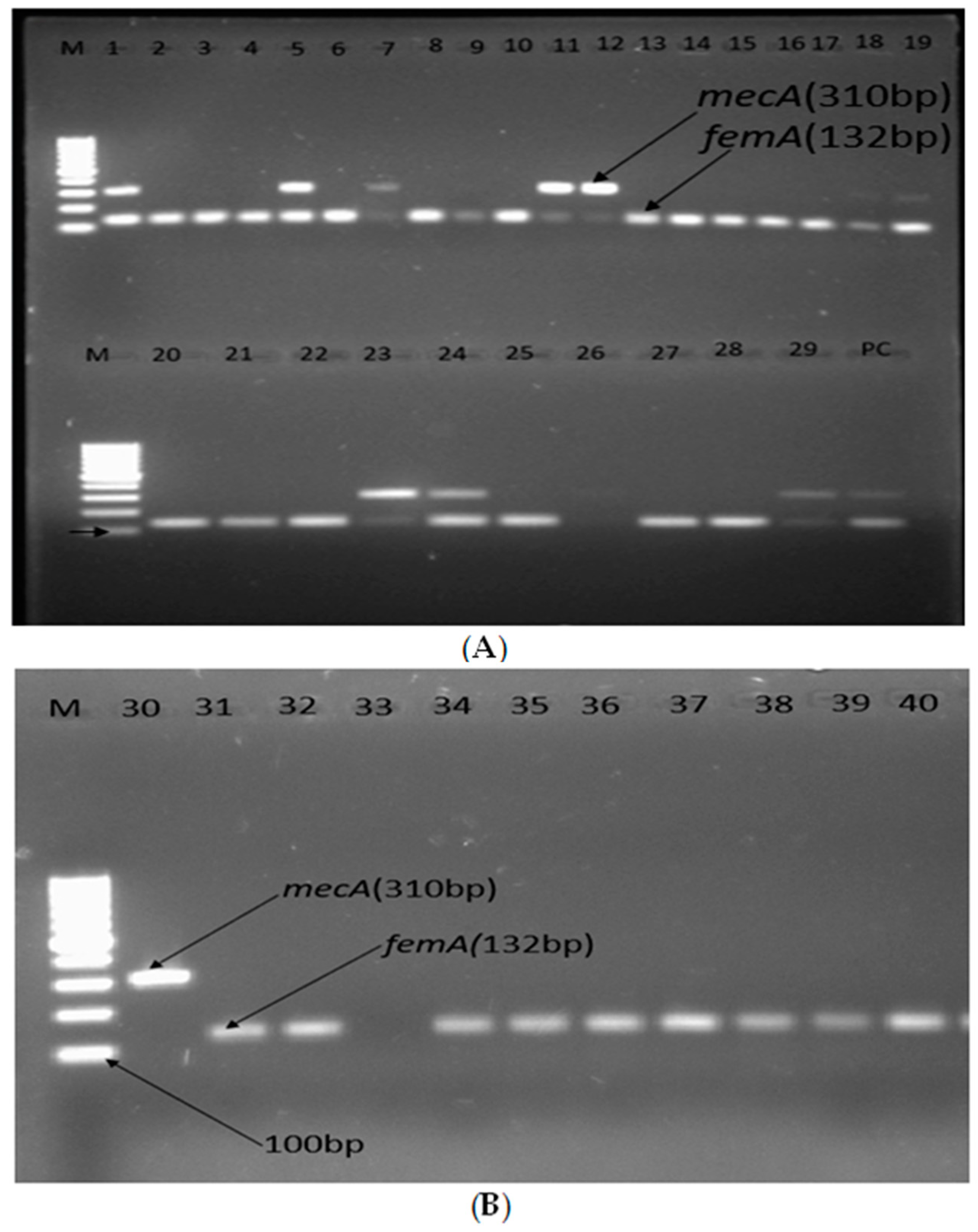
Figure 4. (A,B) Agarose gel electrophoresis showing bands of femA and mecA genes of methicillin-resistant Staphylococcic strains from patients diagnosed with surgical site infection at four different hospitals in Ethiopia; lane M1: 100 bp molecular weight ladder; lane PC: positive control; lanes 1–40 are tested isolates, and positive amplification of femA and mecA is indicated by 132 bp and 310 bp PCR amplicons, respectively.
The 11 isolates that contained the mecA gene, as shown in Figure 4A,B, were S. aureus (lanes 1, 5, 18, and 19), M. sciuri (lanes 7, 23, 24, 29, and 31), S. hemolyticus (lane 11), and S. warneri (lane 12). These were analyzed for vanA and vanB, and none of these isolates showed vanA or vanB in the gel electrophoresis .
Most of the isolates carrying both the mecA and femA gene were reported from Debre Tabor (Figure 5). At Debre Tabor Comprehensive Specialized Hospital, 72.7% of mecA-positive, 70% of cefoxitin-resistant, and 67.7% of femA-positive Staphylococci were discovered (Figure 5).
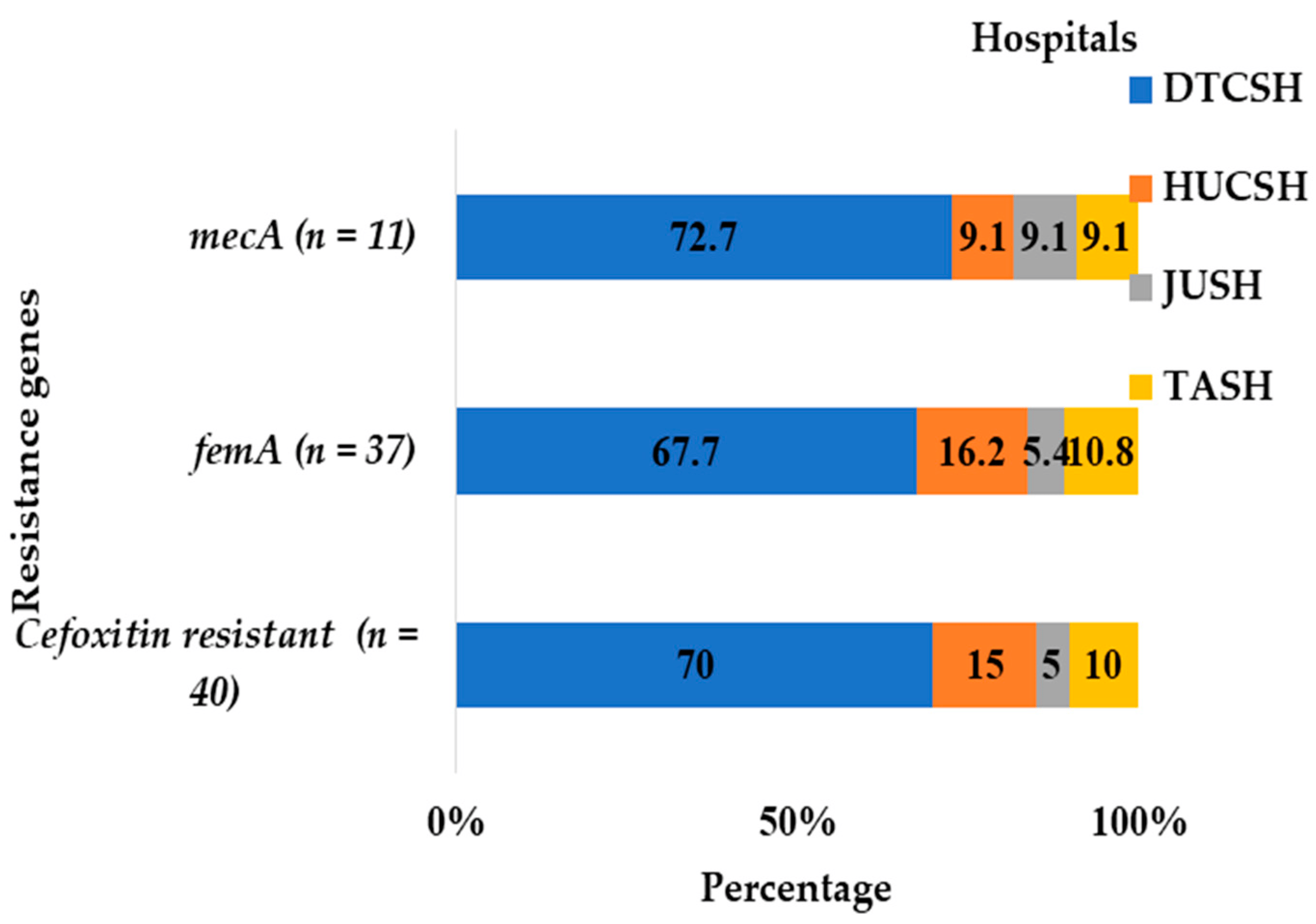
Figure 5. Frequency and distribution of cefoxitin-resistant isolates and mecA and femA genes from the total number of Staphylococci and M. sciuri isolates at each hospital between July 2020 and August 2021. DTCSH: Debre Tabor Comprehensive Specialized Hospital; HUCSH: Hawassa University Comprehensive Specialized Hospital; JUTSH: Jimma University Teaching Specialized Hospital; TASH: Tikur Anbessa Specialized Hospital.
3. Discussion
MRSA is one of the primary bacteria responsible for surgical site infections [18]. The bacteria are human commensals [19], and they can cause a variety of infections, including simple skin and wound infections; they can also infect visceral organs. If not diagnosed and treated properly, many of these illnesses can quickly become life-threatening diseases [1].
In our study, among the 752 wound swab samples processed, we detected 21.7% (163/752) of S. aureus phenotypically. The present finding is similar to previous studies reported from Jimma Ethiopia (23.6%) [20] and Brazil (20%) [21]. On the other hand, this finding is lower than those of studies conducted in other parts of Ethiopia, such as Dessie (34.5%) [22] and Debre Markos (39.7%) [23]. The variation in prevalence between studies might be due to variations in the study subjects, the conducted time, and the method employed for the detection of S. aureus [24].
The proportion of MRSA among the isolates based on disc diffusions was 24.5% (40/163). This study’s findings were similar to a previous study conducted in an Indian Hospital (21.7%) [25]. On the other hand, the finding showed higher frequency than earlier studies in Ethiopia from Dessie (9.8%) [22] and Debre Markos (13.2%) [23], but it was below the national pooled prevalence estimate of Ethiopia (32.5%) [16], Addis Ababa (68.4%) [26], Arba Minch (82.3%) [27], and Nigeria (44%) [28]. Variations in MRSA prevalence across countries are influenced by demographics, antibiotic prescription policies, infection prevention and control programs, staff and elderly hygiene education, healthcare system structure, and MRSA diagnostic facilities [29,30].
From those tested for vancomycin resistance, one isolate had a minimum inhibitory concentration for vancomycin greater than 8 µg/mL, and it was identified as a vancomycin-resistant Staphylococcus. This result was consistent with Pournajaf et al.’s [31] finding that vancomycin resistance was 2.5%, and this figure was lower than that from a review from Ethiopia, where the pooled prevalence of VRSA was 5.3% [16], as well as the findings from Debre Markos (14.1%) [32] and elsewhere (29.4%) [33].
From all methicillin-resistant Staphylococci, the mecA gene was carried by 27.5% of the isolates. This finding was comparable with a study from Nigeria, where 30.5% of the isolates carried the mecA gene [28]. In the present study, 12.9% of S. aureus carried the mecA gene, which is lower than studies reported in Ethiopia (20%) [34], Bangladesh (25%) [35], Nigeria (38%) [28], and Iran (45.1%) [31]. It should be noted that the majority of isolates exhibiting the mecA gene were discovered in Debre Tabor. Eight (72.7%) of the ten mecA-positive isolates were detected at Debre Tabor Hospital. The reason might be poor socioeconomic status, personal demographics, antibiotic prescription practice, and infection control practices, which are associated with increased MRSA infection rates [29,30].
In our present study, the femA gene was detected in all S. aureus isolates, except two cefoxitin-resistant strains (6.7%). This finding was comparable with a study from China [36]. Additionally, in the present study, the femA gene was found in S. haemolyticus, S. warneri, and 83.3% of M. sciuri cefoxitin-resistant strains. On the other hand, neither mecA nor femA were detected in S. epidermidis [36]. The primers used should be specific to S. aureus; therefore, it is somewhat surprising that the two S. aureus lack the gene and that several other non-aureus isolates carry the gene. The explanation could be mutational changes in S. aureus and gene transfer to other species. All of these isolates have been sent for whole genome sequencing, and this matter will be analyzed further when the results are ready.
It is interesting that a significantly higher proportion of CoNS isolates harbour methicillin resistance genes, where 83.3% of M. sciuri and 50% of S. haemolyticus carried the mecA gene. This is in agreement with early reports that CoNS were the most common species in nosocomial infections and exhibit higher antibiotic resistance rates than S. aureus. This may be explained by the high prevalence of methicillin resistance linked with staphylococcal cassette chromosome (SCCmec) elements in CoNS [37], and they are considered a major reservoir of SCCmec [38]. For instance, Berglund et al. described the likely transfer of a type V SCCmec from methicillin-resistant S. haemolyticus to MSSA, thus transforming into MRSA [9,39]. Another study revealed that the mecA homologue in M. sciuri may be an evolutionary precursor to MRSA pathogenic strains, highlighting the main routes of antibiotic resistance gene transfer [40]. Furthermore, the report demonstrated that MSSA become MRSA by acquiring SCCmec from S. epidermidis through horizontal transfer [41]. These accounts suggest that horizontal interspecies transfer of mobile genetic elements could be a crucial element for MRSA global dissemination [39,40,42].
The absence of the mecA and vanA genes in the MRSA and VRSA samples does not imply the absence of resistance, as resistance may be due to other mutations or cassette-containing resistance genes [43]. Globally, resistant staphylococcal isolates lacking the mecA gene show the possibility for additional mechanisms to compete with mecA in the establishment of MRSA [44,45]. MRSA’s resistance against beta-lactams and methicillin is further complicated by its ability to develop resistance to vancomycin through accidental transmission of the vanA gene from Enterococcal strains [46]. Vancomycin is a glycopeptide antibiotic that prevents the formation of the peptidoglycan layer by binding to the peptide precursor. Antibiotic overuse leads to bacterial resistance, thus prompting the search for new antimicrobial strategies [47]. Genomics can identify antibiotic targets, and live non-multiplying bacteria can be targeted for new antibacterials, potentially resulting in new antibacterials that shorten therapy microorganisms, reduce adverse effects, and potentially reduce antibacterial resistance [48]. Preclinical research explores metal uptake via bacterial metallophores [47]. Bacteriophages have been demonstrated to be antibacterial in animals that are susceptible to certain infectious diseases [48].
In the present study, the likelihood of methicillin-resistant staphylococcus SSI increased among patients aged ≥ 61 years (p = 0.025, AOR = 3.729 (1.179–11.791)). Similar findings have been reported in Brazil [21]. Previous studies have suggested that patients on antibiotics (p = 0. 017), who had a previous wound infection (p = 0.006), and with a hospital stay > 72 h showed an association with MRSA infection [32]. Similarly to our finding, previous use of antibiotics (p = 0.025, AOR = 3.066 (1.101–9.392)) and preoperative hospital stays > 7 days (p = 0.000, AOR = 1.856 (0.688–5.311)) demonstrated an association with methicillin-resistant Staphylococci for SSI. Unlike our study, a report by X. Yang et al. [49] showed that long, invasive procedures used in the ICU, such as tracheal intubation and ventilator usage, along with patients with cerebral infarction and other embolisms increase the likelihood of developing MRSA colonization and further infections.
4. Materials and Methods
4.1. Study Area and Design
A cross-sectional study was conducted in four purposively selected University Teaching Hospitals, including Debre Tabor Comprehensive Specialized Hospital (DTCSH), Tikur Anbessa Specialized Hospital (TASH), Hawassa University Teaching Hospital (HUTH), and Jimma University Teaching Hospital (JUTH) in Ethiopia. These hospitals provide a range of services in both outpatient and inpatient units under different wards, such as general surgical, gynecology, obstetric/maternity, and orthopedics, and they all have microbiology laboratories for culture and antimicrobial sensitivity testing. This study was conducted between July 2020 and August 2021.
4.2. Variables
The variables in this study were MRSA and VRSA infections, socio-demographic characteristics, clinical data, and risk factor variables, such as age, sex, surgical site, length of hospital stay, history of hospital admission, previous use of antibiotics, smoking history, alcohol consumption, type and nature of the surgery, type of antimicrobial prophylaxis, history of previous antibiotic use, surgical procedure performed, and duration of the operation.
4.3. Study Population and Sampling
The study population consisted of patients admitted for elective and emergency surgery in general surgery, gynecology/obstetric, and orthopedics wards. All surgical patients, regardless of their age, who underwent surgery during the study period and developed signs and symptoms of surgical site infection (SSI) within 30 days were included in this study. Consent and/or assent was secured from each participant before the commencement of data collection. Patients who developed SSIs after 30 days following the operation, those who refused to participate, patients with infected burn wounds, and those on treatment were excluded from this study.
4.4. Sample Size and Sampling Technique
A total of 752 clinically diagnosed cases of SSI from different wards were enrolled in this study. The sample size was calculated based on a single population proportion sample size estimation formula (n = Z2 P (1 − P)/d2) using a proportion (P) of 20% [50]. As this was a multicenter study, to increase the sample size, a precision (d) of 0.03 was used, where Z stands for Z statistic with a confidence level of 95% and a Z value of 1.96. Considering a 10% non-response rate, the final total sample size was estimated at 752. Enrollments continued until the necessary sample size was achieved, with proportional allocation among the different hospitals based on patient flow.
4.5. Specimen Collection, Isolation, and Identification of S. aureus
Wound swabs or aspirates were collected based on standard operation procedure (SOP). Conventional bacteriological techniques, such as morphological, cultural, and biochemical characterization, were used to identify strains of S. aureus [51]. The specimens were inoculated on blood agar plates (BAP) (Oxoid, UK), and mannitol salt agar (MSA) (Oxoid) and then incubated at 35 °C for 24 h. The S. aureus isolates were identified through Gram staining, catalase and coagulase tests, including golden yellow colonies on MSA, which were considered phenotypic identification tests.
4.6. Identification Confirmation of the Species of Bacteria Strain Using Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry (MALDI-TOF MS)
All phenotypically cefoxitin-resistant S. aureus isolates were re-identified using MALDI-TOF MS [52] at the Clinical Microbiology Department, Uppsala University Hospital in Sweden. A single colony of bacteria from fresh cultures was smeared onto a MALDI-TOF plate, air-dried, treated with formic acid and MALDI matrix solution, and again air-dried before reading. MALDI-TOF identification scores were automatically generated by the system software [53], and isolates with scores of two and above were accepted, while those with scores below 1.7 and flagged red were rejected. Samples with scores between 1.7 and 2 and flagged yellow were re-analyzed.
4.7. Antimicrobial Susceptibility Testing
Antimicrobial susceptibility testing (AST) was carried out using the cefoxitin disc diffusion test, which is a surrogate marker test for oxacillin resistance following the clinical and laboratory standard institute (CLSI) protocol [54], and the minimum inhibitory concentration (MIC) of the vancomycin strip was determined using the E-test method on MHA. The reference strain S. aureus (ATCC® 25923, Seattle, DC, USA) was used as a quality control. Evidence showed that MRSA is a requisite for VISA [55]. Hence, we screened VRSA/VISA from MRSA isolates.
4.8. Identification of Methicillin-Resistant S. aureus Strains
4.9. Quality Assurance
Specimens were collected according to the recommended standard operating procedures (SOPs). The performance of all prepared culture media (BAP and MSA) was also checked by inoculating control strains, S. aureus (ATCC® 25923), for each new batch of agar plates prepared. In addition, the sterility of culture media was checked by incubating 5% of the prepared media at 37 °C for 24–48 h. In addition, reagents for Gram stain and biochemical tests were checked against control strains of S. aureus. The 0.5 McFarland standard was used to standardize the bacterial suspension inoculum density for the susceptibility test. Each MALDI-TOF run also included S. aureus (ATCC® 25923) as a quality control strain. Furthermore, the performance of the antibiotic disks was evaluated using American-type cell culture (ATCC) controls. As such, S. aureus ATCC® 25923 (cefoxitin zone 21–29 mm) and S. aureus ATCC® 43300 (zone ≤ 21 mm) were used as control strains to determine the performance of the cefoxitin disc diffusion test. S. aureus ATCC® 29213 MIC of vancomycin broth value 0.5–2.0 µg/mL was used as a control strain to measure the performance of vancomycin [54].
4.10. Data Entry and Analysis
The data were checked for completeness, missing values, and coding of questionnaires entered into the Research Electronic Data Capture (RED-Cap). A double data entry method was used to ensure the accuracy of the data, and data were analyzed using STATA version 25. Descriptive statistics were used to present antimicrobial susceptibility patterns. Frequencies and cross-tabulations were used to summarize descriptive statistics. Logistic regression was used to study the effect of independent variables on the dependent variables. p-values less than 0.05 were considered statistically significant.
4.11. Ethical Considerations
The Department of Medical Microbiology, Immunology, and Parasitology (DMIP) and the AHRI/ALERT Research Ethics Committee (AAREC) reviewed and approved this study. Institutional review board (IRB) approval was also obtained from Addis Ababa University’s College of Health Sciences, AAUMF03-008/2020. Selected hospitals received a formal letter from the AHRI and DMIP, and each hospital’s medical directors gave their consent. Written consent/assent was taken from each study participant before initiation of the actual data collection.
Patient information was kept confidential by sharing the laboratory results of research participants only with the designated accountable clinicians. Patients who experienced SSIs were managed according to hospital policy. In general, this study was conducted in accordance with the Declaration of Helsinki.
5. Conclusions
A multicenter study identified 11 mecA-positive Staphylococci species, with 36.4% being MRSA, but no VRSA was found among these MRSA. What is more captivating in this study is a significantly high prevalence of mecA carriage among CoNS, suggesting difficulties in the treatment of patients with CoNS infections. Furthermore, this signifies a huge potential of MSSA conversion to MRSA through horizontal gene transfer, which would make things more complicated. In terms of geographic distribution, out of 11 mecA-gene-positive Staphylococci, 8 (72.7%) were detected in DTCSH, with significant variations between hospitals, suggesting that strategies to control methicillin-resistant Staphylococci should be tailored to specific hospitals. The presence of staphylococcal isolates was linked to factors like older age, hospital stay, antibiotic history, and prophylaxis. Prompt prevention and control measures for MRSA-high-risk populations, including strict adherence to infection prevention methods, periodic surveillance, and antibiotic stewardship programs, are crucial for effective treatment and prevention strategies.
References
- Turner, N.A.; Sharma-Kuinkel, B.K.; Maskarinec, S.A.; Eichenberger, E.M.; Shah, P.P.; Carugati, M.; Holland, T.L.; Fowler, V.G. Methicillin-resistant Staphylococcus aureus: An overview of basic and clinical research. Nat. Rev. Microbiol. 2019, 17, 203–218. [Google Scholar] [CrossRef]
- Martínez-Meléndez, A.; Morfín-Otero, R.; Villarreal-Treviño, L.; González-González, G.; Llaca-Díaz, J.; Rodríguez-Noriega, E.; Camacho-Ortíz, A.; Garza-González, E. Staphylococcal cassette chromosome mec (SCCmec) in coagulase negative staphylococci. Med. Univ. 2015, 17, 229–233. [Google Scholar] [CrossRef]
- Beims, H.; Overmann, A.; Fulde, M.; Steinert, M.; Bergmann, S. Isolation of Staphylococcus sciuri from horse skin infection. Open Vet. J. 2016, 6, 242–246. [Google Scholar] [CrossRef]
- Cheung, G.Y.C.; Bae, J.S.; Otto, M. Pathogenicity and virulence of Staphylococcus aureus. Virulence 2021, 12, 547–569. [Google Scholar] [CrossRef] [PubMed]
- Ghssein, G.; Ezzeddine, Z. The Key Element Role of Metallophores in the Pathogenicity and Virulence of Staphylococcus aureus: A Review. Biology 2022, 11, 1525. [Google Scholar] [CrossRef] [PubMed]
- David, M.Z.; Daum, R.S. Treatment of Staphylococcus aureus Infections. Curr. Top. Microbiol. Immunol. 2017, 409, 325–383. [Google Scholar]
- Hiramatsu, K.; Katayama, Y.; Matsuo, M.; Sasaki, T.; Morimoto, Y.; Sekiguchi, A.; Baba, T. Multi-drug-resistant Staphylococcus aureus and future chemotherapy. J. Infect. Chemother. 2014, 20, 593–601. [Google Scholar] [CrossRef] [PubMed]
- Mahjabeen, F.; Saha, U.; Mostafa, M.N.; Siddique, F.; Ahsan, E.; Fathma, S.; Tasnim, A.; Rahman, T.; Faruq, R.; Sakibuzzaman, M.; et al. An Update on Treatment Options for Methicillin-Resistant Staphylococcus aureus (MRSA) Bacteremia: A Systematic Review. Cureus 2022, 14, e31486. [Google Scholar] [CrossRef]
- Vestergaard, M.; Frees, D.; Ingmer, H. Antibiotic Resistance and the MRSA Problem. Microbiol. Spectr. 2019, 7. [Google Scholar] [CrossRef]
- Li, X.; Xiong, Y.; Fan, X.; Feng, P.; Tang, H.; Zhou, T. The role of femA regulating gene on methicillin-resistant Staphylococcus aureus clinical isolates. Med. Mal. Infect. 2012, 42, 218–225. [Google Scholar] [CrossRef]
- Cong, Y.; Yang, S.; Rao, X. Vancomycin resistant Staphylococcus aureus infections: A review of case updating and clinical features. J. Adv. Res. 2020, 21, 169–176. [Google Scholar] [CrossRef]
- Stogios, P.J.; Savchenko, A. Molecular mechanisms of vancomycin resistance. Protein Sci. 2020, 29, 654–669. [Google Scholar] [CrossRef]
- Li, G.; Walker, M.J.; De Oliveira, D.M.P. Vancomycin Resistance in Enterococcus and Staphylococcus aureus. Microorganisms 2022, 11, 24. [Google Scholar] [CrossRef]
- Micek, S.T. Alternatives to vancomycin for the treatment of methicillin-resistant Staphylococcus aureus infections. Clin. Infect. Dis. 2007, 45 (Suppl. S3), S184–S190. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Sabokroo, N.; Wang, Y.; Hashemian, M.; Karamollahi, S.; Kouhsari, E. Systematic review and meta-analysis of the epidemiology of vancomycin-resistance Staphylococcus aureus isolates. Antimicrob. Resist. Infect. Control 2021, 10, 101. [Google Scholar] [CrossRef] [PubMed]
- Eshetie, S.; Tarekegn, F.; Moges, F.; Amsalu, A.; Birhan, W.; Huruy, K. Methicillin resistant Staphylococcus aureus in Ethiopia: A meta-analysis. BMC Infect. Dis. 2016, 16, 689. [Google Scholar] [CrossRef]
- Anagaw, B.; Shiferaw, Y.; Anagaw, B.; Biadglegne, F.; Moges, F.; Kassu, A.; Unakal, C.; Mulu, A. Frequency of methicillin-resistant Staphylococcus aureus isolates from clinical specimens in Gondar University Hospital, Northwest Ethiopia. Asian J. Med. Sci. 2013, 5, 59–64. [Google Scholar] [CrossRef]
- Pal, S.; Sayana, A.; Joshi, A.; Juyal, D. Staphylococcus aureus: A predominant cause of surgical site infections in a rural healthcare setup of Uttarakhand. J. Fam. Med. Prim. Care 2019, 8, 3600–3606. [Google Scholar]
- Haaber, J.; Penadés, J.R.; Ingmer, H. Transfer of antibiotic resistance in Staphylococcus aureus. Trends Microbiol. 2017, 25, 893–905. [Google Scholar] [CrossRef] [PubMed]
- Godebo, G.; Kibru, G.; Tassew, H. Multidrug-resistant bacterial isolates in infected wounds at Jimma University Specialized Hospital, Ethiopia. Ann. Clin. Microbiol. Antimicrob. 2013, 12, 17. [Google Scholar] [CrossRef]
- Almeida, G.C.; dos Santos, M.M.; Lima, N.G.; Cidral, T.A.; Melo, M.C.; Lima, K.C. Prevalence and factors associated with wound colonization by Staphylococcus spp. and Staphylococcus aureus in hospitalized patients in inland northeastern Brazil: A cross-sectional study. BMC Infect. Dis. 2014, 14, 328. [Google Scholar] [CrossRef]
- Tsige, Y.; Tadesse, S.; G/Eyesus, T.; Tefera, M.M.; Amsalu, A.; Menberu, M.A.; Gelaw, B. Prevalence of Methicillin-Resistant Staphylococcus aureus and Associated Risk Factors among Patients with Wound Infection at Referral Hospital, Northeast Ethiopia. J. Pathog. 2020, 2020, 3168325. [Google Scholar] [CrossRef] [PubMed]
- Kahsay, A.; Mihret, A.; Abebe, T.; Andualem, T. Isolation and antimicrobial susceptibility pattern of Staphylococcus aureus in patients with surgical site infection at Debre Markos Referral Hospital, Amhara Region, Ethiopia. Arch. Public Health 2014, 72, 16. [Google Scholar] [CrossRef] [PubMed]
- Congdon, S.T.; Guaglione, J.A.; Ricketts, O.M.A.; Murphy, K.V.; Anderson, M.G.; Trowbridge, D.A.; Abduladheem, Y.A.; Phillips, A.M.; Beausoleil, A.M.; Stanley, A.J.; et al. Prevalence and antibiotic resistance of Staphylococcus aureus associated with a college-aged cohort: Life-style factors that contribute to nasal carriage. Front. Cell. Infect. Microbiol. 2023, 13, 1195758. [Google Scholar] [CrossRef] [PubMed]
- Kownhar, H.; Shankar, E.M.; Vignesh, R.; Sekar, R.; Velu, V.; Rao, U.A. High isolation rate of Staphylococcus aureus from surgical site infections in an Indian hospital. J. Antimicrob. Chemother. 2008, 61, 758–760. [Google Scholar] [CrossRef] [PubMed]
- Tadesse, S.; Alemayehu, H.; Tenna, A.; Tadesse, G.; Tessema, T.S.; Shibeshi, W.; Eguale, T. Antimicrobial resistance profile of Staphylococcus aureus isolated from patients with infection at Tikur Anbessa Specialized Hospital, Addis Ababa, Ethiopia. BMC Pharmacol. Toxicol. 2018, 19, 24. [Google Scholar] [CrossRef] [PubMed]
- Mama, M.; Aklilu, A.; Misgna, K.; Tadesse, M.; Alemayehu, E. Methicillin-and inducible clindamycin-resistant Staphylococcus aureus among patients with wound infection attending Arba Minch Hospital, South Ethiopia. Int. J. Microbiol. 2019, 2019, 2965490. [Google Scholar] [CrossRef] [PubMed]
- Ibadin, E.E.; Enabulele, I.O.; Muinah, F. Prevalence of mecA gene among staphylococci from clinical samples of a tertiary hospital in Benin City, Nigeria. Afr. Health Sci. 2017, 17, 1000–1010. [Google Scholar] [CrossRef]
- Kavanagh, K.T. Control of MSSA and MRSA in the United States: Protocols, policies, risk adjustment and excuses. Antimicrob. Resist. Infect. Control 2019, 8, 103. [Google Scholar] [CrossRef]
- Al-Orphaly, M.; Hadi, H.A.; Eltayeb, F.K.; Al-Hail, H.; Samuel, B.G.; Sultan, A.A.; Skariah, S. Epidemiology of multidrug-resistant Pseudomonas aeruginosa in the Middle East and North Africa Region. mSphere 2021, 6, e00202–e00221. [Google Scholar] [CrossRef]
- Pournajaf, A.; Ardebili, A.; Goudarzi, L.; Khodabandeh, M.; Narimani, T.; Abbaszadeh, H. PCR-based identification of methicillin–resistant Staphylococcus aureus strains and their antibiotic resistance profiles. Asian Pac. J. Trop. Biomed. 2014, 4, S293–S297. [Google Scholar] [CrossRef] [PubMed]
- Tefera, S.; Awoke, T.; Mekonnen, D. Methicillin and vancomycin resistant Staphylococcus aureus and associated factors from surgical ward inpatients at Debre Markos Referral Hospital, Northwest Ethiopia. Infect. Drug Resist. 2021, 14, 3053–3062. [Google Scholar] [CrossRef] [PubMed]
- Alani, H.A.; Hassawi, D.S.; Flayih, M.T. Patterns of antibiotic resistance in Staphylococcus aureus isolates and detection the heteroresistance to vancomycin by population analysis method. JUAPS 2017, 11, 26–33. [Google Scholar] [CrossRef]
- Moges, F.; Tamiru, T.; Amare, A.; Mengistu, G.; Eshetie, S.; Dagnew, M.; Feleke, T.; Gizachew, M.; Abebe, W. Prevalence of Methicillin-Resistant Staphylococcus aureus and Multidrug-Resistant Strains from Patients Attending the Referral Hospitals of Amhara Regional State, Ethiopia. Int. J. Microbiol. 2023, 2023, 3848073. [Google Scholar] [CrossRef]
- Zahan, N.A.; Hossain, M.A.; Musa, A.K.; Shamsuzzaman, A.K.; Mahamud, M.C.; Mamun, A.A.; Paul, S.K.; Ahmed, S.; Sumona, A.A.; Begum, Z.; et al. PCR for mecA gene of methicillin resistant Staphylococcus aureus. Mymensingh Med. J. 2009, 18, 21–26. [Google Scholar] [PubMed]
- Kobayashi, N.; Wu, H.; Kojima, K.; Taniguchi, K.; Urasawa, S.; Uehara, N.; Omizu, Y.; Kishi, Y.; Yagihashi, A.; Kurokawa, I. Detection of mecA, femA, and femB genes in clinical strains of using polymerase chain reaction. Epidemiol. Infect. 1994, 113, 259–266. [Google Scholar] [CrossRef]
- Garza-Gonzalez, E.; Morfin-Otero, R.; Llaca-Diaz, J.M.; Rodriguez-Noriega, E. Staphylococcal cassette chromosome mec (SCCmec) in methicillin-resistant coagulase-negative staphylococci. Epidemiol. Infect. 2010, 138, 645–654. [Google Scholar] [CrossRef]
- Zong, Z.; Peng, C.; Lü, X. Diversity of SCC mec elements in methicillin-resistant coagulase-negative staphylococci clinical isolates. PLoS ONE 2011, 6, e20191. [Google Scholar] [CrossRef]
- Berglund, C.; Söderquist, B. The origin of a methicillin-resistant Staphylococcus aureus isolate at a neonatal ward in Sweden—Possible horizontal transfer of a staphylococcal cassette chromosome mec between methicillin-resistant Staphylococcus haemolyticus and Staphylococcus aureus. Clin. Microbiol. Infect. 2008, 14, 1048–1056. [Google Scholar] [CrossRef]
- Wu, S.W.; de Lencastre, H.; Tomasz, A. Recruitment of the mecA gene homologue of Staphylococcus sciuri into a resistance determinant and expression of the resistant phenotype in Staphylococcus aureus. J. Bacteriol. 2001, 183, 2417–2424. [Google Scholar] [CrossRef]
- Bloemendaal, A.L.A.; Brouwer, E.C.; Fluit, A.C. Methicillin resistance transfer from Staphylocccus epidermidis to methicillin-susceptible Staphylococcus aureus in a patient during antibiotic therapy. PLoS ONE 2010, 5, e11841. [Google Scholar] [CrossRef] [PubMed]
- Miragaia, M. Factors Contributing to the Evolution of mecA-Mediated β-lactam Resistance in Staphylococci: Update and New Insights From Whole Genome Sequencing (WGS). Front. Microbiol. 2018, 9, 2723. [Google Scholar] [CrossRef]
- Sun, J.; Deng, Z.; Yan, A. Bacterial multidrug efflux pumps: Mechanisms, physiology and pharmacological exploitations. Biochem. Biophys. Res. 2014, 453, 254–267. [Google Scholar] [CrossRef] [PubMed]
- Zong, Z. Characterization of a complex context containing mecA but lacking genes encoding cassette chromosome recombinases in Staphylococcus haemolyticus. BMC Microbiol. 2013, 13, 64. [Google Scholar] [CrossRef] [PubMed]
- Elhassan, M.M.; Ozbak, H.A.; Hemeg, H.A.; Elmekki, M.A.; Ahmed, L.M. Absence of the mecA gene in methicillin resistant Staphylococcus aureus isolated from different clinical specimens in Shendi city, Sudan. BioMed. Res. Int. 2015, 2015, 895860. [Google Scholar] [CrossRef] [PubMed]
- McGuinness, W.A.; Malachowa, N.; DeLeo, F.R. Vancomycin Resistance in Staphylococcus aureus. Yale J. Biol. Med. 2017, 90, 269–281. [Google Scholar]
- Ezzeddine, Z.; Ghssein, G. Towards new antibiotics classes targeting bacterial metallophores. Microb. Pathog. 2023, 182, 106221. [Google Scholar] [CrossRef]
- Coates, A.R.; Hu, Y. Novel approaches to developing new antibiotics for bacterial infections. Br. J. Pharmacol. 2007, 152, 1147–1154. [Google Scholar] [CrossRef]
- Yang, X.; Zhao, J.; Wang, Y.; Wu, J.; Wang, X.; Wang, Y.; Zhang, Y.; Li, H. Molecular Epidemiology of Methicillin-Resistant Staphylococcus aureus in Hospitalized Patients in Eastern Heilongjiang Province, China. Infect. Drug Resist. 2021, 14, 1635–1643. [Google Scholar] [CrossRef]
- Mengesha, R.E.; Kasa, B.G.; Saravanan, M.; Berhe, D.F.; Wasihun, A.G. Aerobic bacteria in post surgical wound infections and pattern of their antimicrobial susceptibility in Ayder Teaching and Referral Hospital, Mekelle, Ethiopia. BMC Res. Notes 2014, 7, 575. [Google Scholar] [CrossRef]
- Bendary, M.M.; Solyman, S.M.; Azab, M.M.; Mahmoud, N.F.; Hanora, A.M. Genetic diversity of multidrug resistant Staphylococcus aureus isolated from clinical and non clinical samples in Egypt. Cell. Mol. Biol. 2016, 62, 55–61. [Google Scholar] [PubMed]
- Torres-Sangiao, E.; Leal Rodriguez, C.; García-Riestra, C. Application and Perspectives of MALDI–TOF Mass Spectrometry in Clinical Microbiology Laboratories. Microorganisms 2021, 9, 1539. [Google Scholar] [CrossRef] [PubMed]
- Hou, T.-Y.; Chiang-Ni, C.; Teng, S.-H. Current status of MALDI-TOF mass spectrometry in clinical microbiology. J. Food Drug Anal. 2019, 27, 404–414. [Google Scholar] [CrossRef] [PubMed]
- Clinical Laboratory Standard Institute. Performans Standards for Antimicrobial Susceptablity Testing, CLSI Supplement M100, 30th ed.; Clinical and Laboratory Standared Institiute: Wayne, PA, USA, 2021. [Google Scholar]
- Centers for Disease Control and Prevention. Laboratory Detection of Vancomycin-Intermediate/Resistant Staphylococcus aureus (VISA/VRSA); Centers for Disease Control and Prevention: Atlanta, GA, USA, 2006.
- Yamagishi, J.; Sato, Y.; Shinozaki, N.; Ye, B.; Tsuboi, A.; Nagasaki, M.; Yamashita, R. Comparison of boiling and robotics automation method in DNA extraction for metagenomic sequencing of human oral microbes. PLoS ONE 2016, 11, e0154389. [Google Scholar] [CrossRef]
- Perez-Roth, E.; Claverie-Martın, F.; Villar, J.; Mendez-Alvarez, S. Multiplex PCR for simultaneous identification of Staphylococcus aureus and detection of methicillin and mupirocin resistance. J. Clin. Microbiol. 2001, 39, 4037–4041. [Google Scholar] [CrossRef]
- Emamie, A.; Zolfaghari, P.; Zarei, A.; Ghorbani, M. Prevalence and antibiotic resistance of ESKAPE pathogens isolated from patients with bacteremia in Tehran, Iran. Indian J. Med. Spec. 2023, 14, 97. [Google Scholar]
- Al-Marzoqi, A.; Azize, H.; Al Dulaimi, T.; Ahmed, N. Phenotypic detection of resistance in Staphylococcus aureus isolates: Detection of (mec A and fem A) gene in methicillin resistant Staphylococcus aureus (MRSA) by Polymerase Chain Reaction. J. Nat. Sci. Res. 2014, 4, 112–118. [Google Scholar]

