1. Introduction
Gastropathy is a condition that affects the gastric mucosa [1]. It is characterized by the prevalence of mononuclear infiltrates, the formation of lymphoid follicles with germinal centres in the lamina propria, and subsequent tissue inflammation and regeneration. Histological injury is frequently located in the gastric antrum; however, it sometimes extends to the gastric corpus [2]. Gastropathies are classified according to their aetiologies. Follicular gastropathy (also known as reactive or chemical gastropathy) is mainly attributed to irritants, and their recurrent abuse, including alcohol and nonsteroidal anti-inflammatory drugs (NSAIDs) [1], which play an important role in the development of bacterial infections in the mucosa, particularly those induced by Helicobacter pylori [3]. Approximately 50% of the worldwide population is colonized with H. pylori, representing up to 90% of the gastric microbiota of healthy subjects [4]. However, only 1–2% of this population develops several degrees of chronic mucosal inflammation with clinical manifestations and severe complications [5]. The Mexican consensus on the diagnosis, prevention, and treatment of NSAID-induced gastropathy and enteropathy [6] has determined that the consumption of NSAIDs in subjects with H. pylori infection is a risk factor (relative risk = 20.8) for the development of gastroduodenopathies [7].
H. pylori is classified as a high-priority pathogen because of its resistance to multiple antibiotics [8]. Its association with several infectious diseases has led us to seriously consider the most accurate diagnoses for the optimal selection of treatment therapies. Several non-invasive diagnostic techniques, such as the urea breath test, blood and stool tests, and upper gastrointestinal series [9], have been widely developed for the detection of this bacterium. However, endoscopic studies, which are considered invasive methods for clinical diagnosis, remain helpful because they facilitate the early identification of the aetiological agent [10].
H. pylori eradication has been suggested in patients with gastropathy, who are undergoing NSAIDs therapy, to avoid subsequent complications (i.e., development of ulcerations) [6]. International guidelines for the treatment of H. pylori infections suggest the use of the standard triple therapy (STT), which is considered the first therapeutic alternative for effective eradication [11]. This therapy consists of two antibiotics and a proton pump inhibitor (PPI), usually composed of clarithromycin (CLR), amoxicillin (AMX), and omeprazole [12]. However, clinical guides for the eradication of H. pylori in Mexico and several European countries have replaced the use of CLR by considering metronidazole (MTZ) as a first-line drug for treating this infection [13,14].
Antibiotics have improved the outcomes of treatment therapies for bacterial infectious diseases. They are considered one of the major contributors to the rise in life expectancy by exponentially diminishing morbidity and mortality rates. Unfortunately, the sometimes-unjustified employment and abuse of drugs for the relief of gastric symptomatology or self-treatment of several infectious diseases have contributed to the global crisis of resistance to multiple antibiotics. Although antibiotics are widely employed for the treatment of specific bacterial infections [15], they can cause several states of dysbiosis.
Dysbiosis, defined as the induction of several alterations in the native microbiota of the host, can be observed as a significant reduction or permanent loss of commensal species in the affected microbial communities and/or the establishment, colonization, or overgrowth of gastrointestinal pathobionts (native commensal microbiota that presents pathogen behaviours under dysbiotic conditions) [16]. Although H. pylori induces dysbiosis in infected patients by colonizing up to 90% of the total gastric microenvironment for its establishment, recent studies have highlighted the possible association of non-H. pylori bacteria with the initiation and development of not only follicular gastropathy and pangastropathy but also several gastric infectious diseases [17] by correlating bacterial overgrowth under non-favourable microenvironmental conditions [18], such as the pathobiont Cutibacterium acnes.
The species of the genus Cutibacterium reside as dominant commensal bacteria in the human skin [19]. Although most Cutibacterium species are adapted to inhabit the human skin [20,21], C. acnes is mainly associated with the maintenance of skin homeostasis due to its multiple benefits in this organ. This species was first reported in healthy gastric mucosa [22,23,24]; however, its functions in the gastric microenvironment have not been fully elucidated. C. acnes has recently been classified as a possible trigger of gastric clinical outcomes, such as corpus-dominant lymphocytic gastritis [25], and has been considered a high-risk factor for the initiation and development of gastric cancer [26].
This study aimed to characterize the gastric microbiota of patients with follicular gastropathy and pangastropathy, evaluate the effects of STT on the gastric environment, and correlate the presence of pathobiont C. acnes with the development of gastric infectious diseases.
2. Materials and Methods
2.1. Characterization of the Gastric Microenvironment
2.2. Correlation of the Presence of C. acnes and H. pylori with Gastric Diseases
3. Results
3.1. Characterization of the Gastric Microenvironment
3.2. Correlation of C. acnes in Gastric Diseases
Forty FFPE gastric biopsy samples from patients with non-significant histologic alterations, gastritis, gastropathies, and intestinal metaplasia were evaluated using qRT-PCR. Figure 7 shows the correlation between the presence of C. acnes and H. pylori in the FFPE samples obtained from patients with gastric infectious diseases. Seventeen patients revealed C. acnes transcripts, whereas H. pylori was determined as absent. Five patients with non-significant alterations were exclusively colonized with C. acnes. However, when evaluating gastropathies, six samples were presented with both bacteria. The number of C. acnes transcripts was significantly higher than that of H. pylori transcripts. Finally, when evaluating the samples from patients diagnosed with intestinal metaplasia, only one patient presented with C. acnes transcripts. The remaining patients did not present C. acnes or H. pylori transcripts .
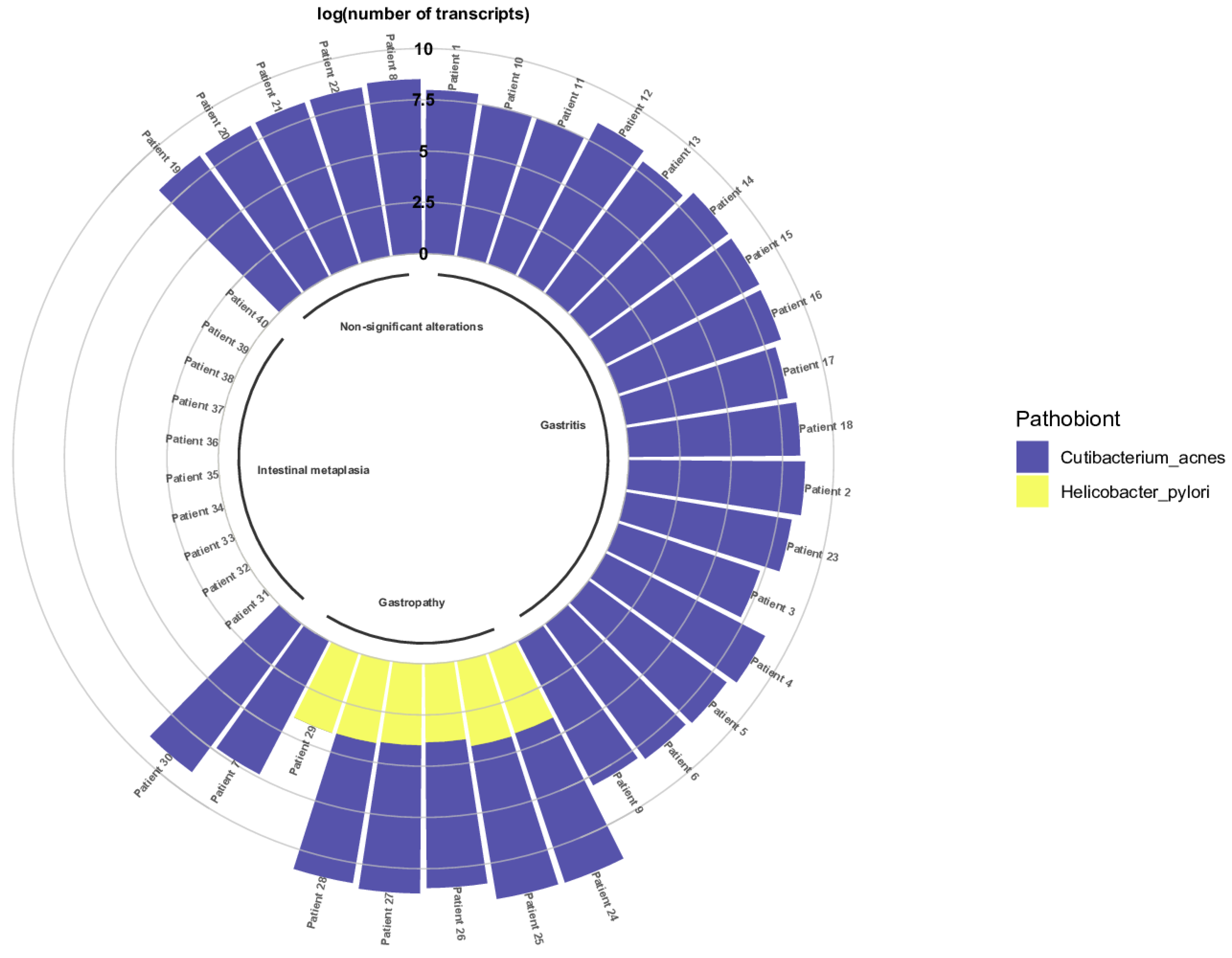
Figure 7. Presence of C. acnes and H. pylori in patients with gastric diseases. Evaluation of the presence of C. acnes and H. pylori in FFPE gastric biopsies via the determination of the log of the number of transcripts of both bacteria in the gastric biopsy samples.
As observed in , the number of C. acnes transcripts was determined as statistically significant compared with the number of H. pylori transcripts, with the non-parametric Mann–Whitney U-Test.
The number of C. acnes transcripts was determined significantly higher than the number of H. pylori transcripts .
Additionally, the significance of the number of C. acnes transcripts by pathology was determined using the Kruskal–Wallis test . As observed, the number of C. acnes transcripts was higher in gastritis (maximum = 573,263,008.44), whereas it was lower in gastropathies (maximum = 48,151,777.41).
4. Discussion
The health status of the host can be significantly altered by the induction of dysbiosis in the human gastrointestinal microbiome, which increases the susceptibility to gastric disorders [16], and difficulting both clinical diagnosis and treatment of infections.
The diagnosis for the identification of H. pylori infection is not easy [36]. Therefore, different methodologies are necessary for clinical diagnosis. Nevertheless, each technique presents variable sensitivity and specificity. Histopathological evaluation revealed that patients 3B and 4B tested positive for H. pylori. However, microbiological and molecular analyses only confirmed patients 1B, 5B, and 6B as H. pylori positive. Metagenomics revealed H. pylori representative sequences in patients 3B (interpreted as an occult infection event), 5B, and 6B, but not in patient 1B. Although this may be interpreted as a false-negative result, we attribute this result to the sampling site. Conflicting reports regarding the best sampling site for the detection of H. pylori continue because of the migration mechanisms of this bacterium in the stomach [37,38]. Advanced stages of histological injury have been also implied in the diminishment of H. pylori populations, reducing its detection rates [39] and hindering its identification using molecular methods, including metagenomics.
Coccoid structures were detected in patient 5A via histopathological evaluation. Although H. pylori can modify its native morphology under dysbiotic microenvironmental conditions (i.e., antibiotic and antisecretory therapies, and accumulation of toxigenic metabolic products (reactive O2 species, pyrimidine nucleotides)) as an adaptative mechanism [40,41], it was impossible to conclude its presence. Therefore, we attribute this finding to the muco-microbiotic layer, which is described from a morphofunctional and histological point of view, as the first line of defense under hostile conditions. This structure is the product of the union of the mucus layer and microorganisms. Although in most routine histologic evaluations the muco-microbiotic layer is unfortunately not visible due to the processing of the sample, the detection of microbial morphologies in H. pylori-negative subjects, as observed in our study, can elucidate a possible key role of these bacteria in the gastrointestinal physiology and pathophysiology [42].
Patients 1B, 6B, and 4B presented with urealytic activity. Although the urea breath test is one of the fastest and most employed tests for clinical diagnosis [43], studies have reported false-positive rates of up to 16.9% [44,45], supporting other reports that have also observed native commensal bacteria with urealytic activity in the stomach, e.g., Staphylococcus epidermidis, Streptococcus salivarius, and Staphylococcus capitis urealiticum [46].
Although these results strongly suggest a lack of trustworthy in the different tests for clinical diagnosis, this study attempts to highlight the importance of the integration of all diagnostic tools for the identification of H. pylori. We suggest not treating all results as mutually excluding but as complementary data.
Although the international guidelines for the treatment of H. pylori infection usually suggest the use of the STT composed of CLR, AMX, and omeprazole [8,9], this therapy has lost its effectiveness due to the increasing resistance rates to antibiotics [47], lengthened periods of treatment up to 2 weeks, and the intention to reduce treatment periods to less than 1 week [48]. To date, STT with CLR is considered one of the least effective treatments since its eradication rates stand < 73% [49], forcing the clinical practice to reconsider other broad-spectrum antibiotics, such as MTZ, which has shown an improvement in the symptomatology and eradication rates > 94.3% when combined with AMX and omeprazole [48].
The lack of objectivity in selecting an optimal therapy according to the phenotypic and genotypic characteristics of H. pylori remains a major challenge, particularly with regard to the increasing rates of antimicrobial resistance, which in consequence hamper the treatment of the bacterial infection, and rapidly increases the gastroduodenal morbidity rates [50]. Three primary cultures isolated from the pretreatment patients presented with specific susceptibility profiles to the four antibiotics. However, when testing the subcultures, discrepancies were observed in their susceptibility profiles. Although the emergence of multidrug-resistant strains has been recognized as a growing problem for the treatment of these infections, heteroresistance, a non-widely discussed issue, must be examined [50].
Heteroresistance, which is defined as the different susceptibility profiles to specific antibiotics in H. pylori subpopulations [51], has been poorly detected mainly due to the lack of standardized methods for its characterization. However, our findings justify the need for new methods for the optimal detection and genotyping of these subpopulations prior to a multiple-antibiotic therapy prescription [50].
Clinical outcomes are attributed to strain-specific virulence factors [52]. Although cagA+ increases the risk of mucosal inflammation [53,54], highly diverse vacA s1m1/s2m2 variants were correlated with more severe histological lesions and clinical outcomes [55,56]. Demirturk et al. [57] reported severe atrophy in diverse cagA and vacA genotypes, revealing a higher risk of progression of precancerous lesions than each virulence factor considered separately. Urtiz-Estrada et al. [58] identified that the cagA+vacAs1+m1 genotype is the most prevalent genotype in Mexican patients and is correlated with various gastric diseases. In our study, it was the most observed genotype; however, some studies widely suggest the possible association of non-H. pylori microbiota with the induction of histological injuries in H. pylori-negative patients [25].
Patient 2A revealed H. pylori representative sequences after the administration of STT. Buffie et al. [59] reported that dysbiotic events can facilitate the establishment of infections as recent acquisition events during the recolonization period, which was determined after the administration of STT. Although some studies reported de novo infections via the inadequate sterilization of surgical materials [60,61], other factors must be considered for these types of events, such as the habits of the host during the infection treatment process, including age, sex, diet, fomites, lipid metabolism, smoking, alcohol consumption, and physical activity [62].
Although dysbiosis is mainly attributed to STT administration, the broad-spectrum activity of the antibiotics prevented the development of infections by not allowing exogenous bacteria to infiltrate the mucosal tissue during therapy, in addition to the competition mechanisms for the inhibition of colonization resistance, pressure selection, and regulation of overgrowth by the survivor gastric microbiota (production of bacteriocins, alterations in gastric pH, consumption of limited resources for competition, and promotion of the epithelial barrier by antimicrobial peptides) [63,64].
Bacterial communities that were altered during dysbiosis (Figure 7) have been reported to be involved in the maintenance of gastrointestinal homeostasis, e.g., signalling for the release of gastric acids [65]; regulation of pathobiont overgrowth [66]; synthesis of precursors in the synthesis of short-chain fatty acids [67]; metabolism of processed foods and production of histamine under halophilic conditions [68]; immunomodulation [69,70,71,72]; and lipid digestion processes [73]. Some of these bacteria have also been shown to be associated with secondary infections in immunocompromised patients [74,75,76,77,78,79,80].
Diversity and richness index values after the recolonization period varied between individuals. The overgrowth and dominance of pathobiont bacteria of clinical interest were observed; however, both metrics after the administration of STT slightly increased, suggesting recolonization which we attribute to the lifestyle [16]. Recolonization by pathobionts was observed. Palleja et al. [81] observed recolonization of the intestinal microbiota after the administration of a multiple-antibiotic therapy. Most species recovered their almost native relative abundances 42 days after treatment, suggesting the modulation of recovery patterns by antibiotics resistance genes (ARGs). In our study, recolonization was observed to be predominantly performed by non-dominant facultative bacteria on day 30. Birg, Ritz, and Lin. [64] reported that eradication treatments generate organ-specific dysbiosis by inducing oxygenation of the gastric tissue via severe inflammation processes, favouring the growth of facultative anaerobic pathogens with antibiotic resistance genes (ARGs).
Recolonization by pathobiont bacteria is an actual concern due to their ability to infect patients with an increased risk of infection. Our study revealed the dominance of Pseudomonas after dysbiosis over time [82]. Although this genus colonizes healthy subjects, it can overgrow on almost any surface due to its non-restrictive metabolic requirements. Pseudomonas aeruginosa and Pseudomonas fluorescens are considered to be of clinical interest because of their role as opportunist pathogens in healthcare-associated infections (HAIs) [83,84], in addition to the risk of establishment of carbapenem-resistant P. aeruginosa [82].
Microbial communities with specific functions (the inhibition of H. pylori growth and its conversion to coccoid structures via the modulation of uremic toxins [18]; acquisition and competition of nutrients to prevent the establishment of Escherichia coli pathotypes [85,86,87,88]; bioeradication and recovery from infectious diseases [89,90]; and generation of energy [91,92]) increased their relative abundances in the gastric microenvironment of the study subjects. However, these bacteria have been also associated with immunocompromised patients and those with gastric diseases [85,86,87,88,91,92,93,94,95].
According to the results of our study, we strongly suggest that dysbiotic events, such as bacterial eradication by the administration of a multiple-antibiotic treatment, and its consequences, e.g., survival and overgrowth of adapted microbial communities, indicate a true ecological opportunity for these pathobionts to persist in the microenvironment by benefitting of the alterations induced in the native microbiota, e.g., eradication of non-resistant native microbiota, whose principal functions might include the regulation of the establishment or overgrowth of specific pathobionts or exogenous microbiota, both of clinical interest.
All pre-treatment patients were initially diagnosed with H. pylori infection. Histological injury was observed as well. However, only three patients showed relative abundances < 9% and <1% of this bacterium. Both presence and bacterial overgrowth are widely correlated with the pathogenesis of infectious diseases, which were not observed for H. pylori in this study. Through metagenomics, C. acnes was observed to be dominant in almost all pre-treatment patients, regardless of the presence–absence of histological injury (i.e., patient 2), strongly suggesting the role of non-H. pylori microbiota in not only development but also the initiation of gastric infectious diseases [96].
To support our findings, FFPE gastric biopsy samples were evaluated. H. pylori was exclusively present in samples from patients with gastropathies. This bacterium is the most frequent cause of gastroduodenal diseases due to its evident dominant relative abundances in subjects with these conditions [97]. However, H. pylori has coevolved within humans through time as a pathobiont by inducing multiple benefits within the host [98]. Additionally, studies have reported the incidence of gastric alterations in the absence of this bacterium in worldwide populations, in which the aetiology of the cases could not be determined [99,100], suggesting a possible role of specific gastric bacteria under dysbiosis.
Research on the microbiome and its significant findings regarding the dysbiosis of several clinical outcomes opens the opportunity to study other bacterial agents possibly involved in the initiation and development of infectious diseases [101]. Studies have reported an overgrowth of Paludibacter sp. and Dialister sp. in patients with gastritis [102], whereas other authors have observed the role of an overabundance of Streptococcus spp., Haemophilus parainfluenzae, and Treponema spp. in the development and progression of dysbiosis in patients with non-H. pylori gastritis [103], suggesting new potential associations between the absence of this pathobiont and the development of infectious diseases [101]. Native commensal bacteria (i.e., Lactobacillus spp., Prevotella melaninogenica, Streptococcus anginosus) have also been shown to be associated with the development of peptic ulcer and gastric cancer [17,18]. The skin pathobiont Cutibacterium acnes was present in almost all FFPE samples.
Neither H. pylori nor C. acnes transcripts were identified in intestinal metaplasia gastric biopsy samples, except in one patient. Studies have determined significant differences in gastric microbial diversity, which is gradually reduced while progressing from non-atrophic gastritis to gastric cancer [104] due to an increase in the production of proinflammatory cytokines and subsequent progressive inflammation [105]. Although these conditions result in an inhospitable microenvironment for most native bacteria, studies have reported dysregulation of the bacterial overgrowth of lactic acid bacteria, which can also promote the development of neoplasia [101,105]. Patient 30 was colonized with C. acnes, which has shown overabundance in gastric tumoral tissues because of its ability to induce inflammation via the production of interleukin-15 [105].
5. Conclusions
NGS tools enable us to determine alterations in the microbiota under specific health-disease conditions. The dysbiotic events of specific pathobionts were highlighted by the dominance of C. acnes in the gastric microenvironment, suggesting the possible role of this bacterium in the initiation or development of diseases. However, some limitations could not allow us to conclude its possible role in the stomach. To support our findings, the presence of C. acnes was evaluated, allowing us to highlight its dominance in different gastric alterations. Therefore, gastroduodenal disorders should no longer be considered as self-limiting diseases. Studies regarding the characterization of C. acnes must be conducted to evaluate the functions of this bacterium in the gastric microenvironment and determine its role in the pathogenesis of infectious diseases. This study takes part in the list of studies that have characterized dysbiotic events, in addition to the recent emergence of pathobiont bacterial species, which under a disequilibrium state can induce severe injury to the host, instead of generating multiple benefits to a specific organ.
References
- Zhang, S.L.; Lollie, T.K.; Chen, Z.; Narasimhalu, T.; Wang, H.L. Histopathologic diagnosis of gastritis and gastropathy: A narrative review. Dig. Med. Res. 2022, 6, 3. [Google Scholar] [CrossRef]
- Carlosama-Rosero, Y.; Bolaños-Bravo, H.; Sierra-Tórres, C.; Rosero, E. Asociación de los genotipos cagA, vacA e IceA de H. pylori con la gastritis crónica y folicular en una población colombiana con alto riesgo de cáncer gástrico. Rev. Gastroenterol. Mex. 2019, 84, 158–164. [Google Scholar] [CrossRef] [PubMed]
- Kayacetin, S.; Guresci, S. What is gastritis? What is gastropathy? How is it classified? Turk. J. Gastroenterol. Off. J. Turk. Soc. Gastroenterol. 2014, 25, 233–247. [Google Scholar] [CrossRef] [PubMed]
- Atherton, J.C.; Blaser, M.J. Coadaptation of Helicobacter pylori and humans: Ancient history, modern implications. J. Clin. Investig. 2009, 119, 2475–2487. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.-C. Treatment of Helicobacter pylori infection: Current status and future concepts. World J. Gastroenterol. 2014, 20, 5283–5293. [Google Scholar] [CrossRef]
- Bielsa-Fernández, M.; la Cuesta, J.T.-D.; Lizárraga-López, J.; Remes-Troche, J.; Carmona-Sánchez, R.; Aldana-Ledesma, J.; Avendaño-Reyes, J.; Ballesteros-Amozorrutia, M.; De Ariño, M.; de Giau-Triulzi, L.; et al. Consenso mexicano sobre diagnóstico, prevención y tratamiento de la gastropatía y enteropatía por antiinflamatorios no esteroideos. Rev. Gastroenterol. Mex. 2020, 85, 190–206. [Google Scholar] [CrossRef]
- Lanza, F.L.; Chan, F.K.; Quigley, E.M.; Practice Parameters Committee of the American College of Gastroenterology. Guidelines for Prevention of NSAID-Related Ulcer Complications. Am. J. Gastroenterol. 2009, 104, 728–738. [Google Scholar]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- National Institute of Diabetes and Digestive and Kidney Diseases. Gastritis & Gastropathy. Available online: https://www.niddk.nih.gov/health-information/digestive-diseases/gastritis-gastropathy (accessed on 12 October 2023).
- Kato, T.; Yagi, N.; Kamada, T.; Shimbo, T.; Watanabe, H.; Ida, K.; Study Group for Establishing Endoscopic Diagnosis of Chronic Gastritis. Diagnosis of Helicobacter Pylori Infection in Gastric Mucosa by Endoscopic Features: A Multicenter Prospective Study. Dig. Endosc. Off. J. Jpn. Gastroenterol. Endosc. Soc. 2013, 25, 508–518. [Google Scholar] [CrossRef]
- Malfertheiner, P.; Megraud, F.; Rokkas, T.; Gisbert, J.P.; Liou, J.-M.; Schulz, C.; Gasbarrini, A.; Hunt, R.H.; Leja, M.; O’Morain, C.; et al. Management of Helicobacter pylori infection: The Maastricht VI/Florence consensus report. Gut 2022, 71, 1724–1762. [Google Scholar] [CrossRef]
- Ladrón-De-Guevara, L.; Bornstein-Quevedo, L.; González-Huezo, S.; Castañeda-Romero, B.; Costa, F.; di Silvio-López, M. Erradicación de Helicobacter pylori en México con un esquema basado en levofloxacina versus la triple terapia estándar: Resultados de un estudio clínico de fase iiib, abierto, aleatorizado, de no inferioridad. Rev. Gastroenterol. Mex. 2018, 84, 274–283. [Google Scholar] [CrossRef] [PubMed]
- Guía de Práctica Clínica Diagnóstico y Tratamiento de la Dispepsia Funcional. 2009. México. Instituto Mexicano del Seguro Social. Available online: http://www.imss.gob.mx/sites/all/statics/guiasclinicas/071GER.pdf (accessed on 15 October 2022).
- Wasielica-Berger, J.; Gugnacki, P.; Mlynarczyk, M.; Rogalski, P.; Swidnicka-Siergiejko, A.; Antonowicz, S.; Krzyzak, M.; Maslach, D.; Dabrowski, A.; Daniluk, J. Comparative Effectiveness of Various Eradication Regimens for Helicobacter Pylori Infection in the Northeastern Region of Poland. Int. J. Environ. Res. Public Health 2022, 19, 6921. [Google Scholar] [CrossRef]
- Patangia, D.V.; Ryan, C.A.; Dempsey, E.; Ross, R.P.; Stanton, C. Impact of antibiotics on the human microbiome and consequences for host health. Microbiologyopen 2022, 11, e1260. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Ramirez, U.; Valencia-Mayoral, P.; Mendoza-Elizalde, S.; Murillo-Eliosa, J.R.; Santos, F.S.; Contreras-Rodríguez, A.; Zúñiga, G.; Aguilar-Rodea, P.; Jiménez-Rojas, V.L.; Galindo, J.C.V.; et al. Role of Helicobacter pylori and Other Environmental Factors in the Development of Gastric Dysbiosis. Pathogens 2021, 10, 1203. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Honggang, Y. The Role of Non-H. Pylori Bacteria in the Development of Gastric Cancer. Am. J. Cancer Res. 2020, 10, 2271–2281. [Google Scholar] [PubMed]
- Petra, C.V.; Rus, A.; Dumitrașcu, D.L. Gastric Microbiota: Tracing the Culprit. Med. Pharm. Rep. 2017, 90, 369–376. [Google Scholar] [CrossRef]
- Boisrenoult, P. Cutibacterium acnes prosthetic joint infection: Diagnosis and treatment. Orthop. Traumatol. Surg. Res. 2018, 104, S19–S24. [Google Scholar] [CrossRef]
- Claesen, J.; Spagnolo, J.B.; Ramos, S.F.; Kurita, K.L.; Byrd, A.L.; Aksenov, A.A.; Melnik, A.V.; Wong, W.R.; Wang, S.; Hernandez, R.D.; et al. A Cutibacterium acnes antibiotic modulates human skin microbiota composition in hair follicles. Sci. Transl. Med. 2020, 12, eaay5445. [Google Scholar] [CrossRef]
- Fournière, M.; Latire, T.; Souak, D.; Feuilloley, M.G.J.; Bedoux, G. Staphylococcus epidermidis and Cutibacterium acnes: Two Major Sentinels of Skin Microbiota and the Influence of Cosmetics. Microorganisms 2020, 8, 1752. [Google Scholar] [CrossRef]
- Delgado, S.; Suárez, A.; Mayo, B. Identification, typing and characterisation of Propionibacterium strains from healthy mucosa of the human stomach. Int. J. Food Microbiol. 2011, 149, 65–72. [Google Scholar] [CrossRef]
- Grice, E.A.; Segre, J.A. The skin microbiome. Nat. Rev. Microbiol. 2011, 9, 244–253. [Google Scholar] [CrossRef] [PubMed]
- Brüggemann, H.; Salar-Vidal, L.; Gollnick, H.P.M.; Lood, R. A Janus-Faced Bacterium: Host-Beneficial and -Detrimental Roles of Cutibacterium acnes. Front. Microbiol. 2021, 12, 673845. [Google Scholar] [CrossRef] [PubMed]
- Montalban-Arques, A.; Wurm, P.; Trajanoski, S.; Schauer, S.; Kienesberger, S.; Halwachs, B.; Högenauer, C.; Langner, C.; Gorkiewicz, G. Propionibacterium acnes overabundance and natural killer group 2 member D system activation in corpus-dominant lymphocytic gastritis. J. Pathol. 2016, 240, 425–436. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Shao, L.; Liu, X.; Ji, F.; Mei, Y.; Cheng, Y.; Liu, F.; Yan, C.; Li, L.; Ling, Z. Alterations of gastric mucosal microbiota across different stomach microhabitats in a cohort of 276 patients with gastric cancer. EBioMedicine 2018, 40, 336–348. [Google Scholar] [CrossRef] [PubMed]
- Dixon, M.F.; Genta, R.M.; Yardley, J.H.; Correa, P. Classification and Grading of Gastritis. The updated Sydney System. International Workshop on the Histopathology of Gastritis, Houston 1994. Am. J. Surg. Pathol. 1996, 20, 1161–1181. [Google Scholar] [CrossRef]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 31st ed.; Wayne, P.A., Ed.; CLSI Supplement M100; Clinical and Laboratory Standards Institute: Malvern, PA, USA, 2015. [Google Scholar]
- Mendoza-Elizalde, S.; Arteaga-Resendiz, N.K.; Valencia-Mayoral, P.; Luna, R.C.; Moreno-Espinosa, S.; Arenas-Huertero, F.; Zúñiga, G.; Velázquez-Guadarrama, N. Diversification of the vacAs1m1 and vacAs2m2 Strains of Helicobacter pylori in Meriones unguiculatus. Front. Microbiol. 2016, 7, 1758. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2022; Available online: https://www.r-project.org/ (accessed on 15 October 2022).
- Miura, Y.; Ishige, I.; Soejima, N.; Suzuki, Y.; Uchida, K.; Kawana, S.; Eishi, Y. Quantitative PCR of Propionibacterium acnes DNA in samples aspirated from sebaceous follicles on the normal skin of subjects with or without acne. J. Med. Dent. Sci. 2010, 57, 65–74. [Google Scholar]
- Roshdy, T.M.; Saleh, S.A.; Abass, N.H.; Bassiouny, K.; Khalil, H. Disturbing intracellular replication of Helicobacter pylori by sorafenib treatment in-vitro. Afr. J. Biol. Sci. 2021, 17, 285–295. [Google Scholar] [CrossRef]
- Abadi, A.T.B. Diagnosis of Helicobacter pylori Using Invasive and Noninvasive Approaches. J. Pathog. 2018, 2018, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Ailloud, F.; Didelot, X.; Woltemate, S.; Pfaffinger, G.; Overmann, J.; Bader, R.C.; Schulz, C.; Malfertheiner, P.; Suerbaum, S. Within-host evolution of Helicobacter pylori shaped by niche-specific adaptation, intragastric migrations and selective sweeps. Nat. Commun. 2019, 10, 2273. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.G.; Choi, I.J.; Lee, J.Y.; Cho, S.-J.; Nam, B.-H.; Kook, M.-C.; Hong, E.K.; Kim, Y.-W. Biopsy site for detecting Helicobacter pylori infection in patients with gastric cancer. J. Gastroenterol. Hepatol. 2009, 24, 469–474. [Google Scholar] [CrossRef]
- Noh, C.-K.; Lee, G.H.; Park, J.W.; Roh, J.; Han, J.H.; Lee, E.; Park, B.; Lim, S.G.; Shin, S.J.; Cheong, J.Y.; et al. Diagnostic accuracy of “sweeping” method compared to conventional sampling in rapid urease test for Helicobacter pylori detection in atrophic mucosa. Sci. Rep. 2020, 10, 18483. [Google Scholar] [CrossRef]
- Ierardi, E.; Losurdo, G.; Mileti, A.; Paolillo, R.; Giorgio, F.; Principi, M.; Di Leo, A. The Puzzle of Coccoid Forms of Helicobacter pylori: Beyond Basic Science. Antibiotics 2020, 9, 293. [Google Scholar] [CrossRef]
- Pennelli, G.; Grillo, F.; Galuppini, F.; Ingravallo, G.; Pilozzi, E.; Rugge, M.; Fiocca, R.; Fassan, M.; Mastracci, L. Gastritis: Update on etiological features and histological practical approach. Pathologica 2020, 112, 153–165. [Google Scholar] [CrossRef]
- Fucarino, A.; Burgio, S.; Paladino, L.; Bavisotto, C.C.; Pitruzzella, A.; Bucchieri, F.; Cappello, F. The Microbiota Is Not an Organ: Introducing the Muco-Microbiotic Layer as a Novel Morphofunctional Structure. Anatomia 2022, 1, 186–203. [Google Scholar] [CrossRef]
- Nakamura, H.; Yoshiyama, H.; Takeuchi, H.; Mizote, T.; Okita, K.; Nakazawa, T. Urease Plays an Important Role in the Chemotactic Motility of Helicobacter pylori in a Viscous Environment. Infect. Immun. 1998, 66, 4832–4837. [Google Scholar] [CrossRef]
- Brandi, G.; Biavati, B.; Calabrese, C.; Granata, M.; Nannetti, A.; Mattarelli, P.; Di Febo, G.; Saccoccio, G.; Biasco, G. Urease-Positive Bacteria Other than Helicobacter pylori in Human Gastric Juice and Mucosa. Am. J. Gastroenterol. 2006, 101, 1756–1761. [Google Scholar] [CrossRef]
- Ramírez-Lázaro, M.J.; Lario, S.; Calvet, X.; Sánchez-Delgado, J.; Montserrat, A.; Quílez, E.M.; Casalots, A.; Suarez, D.; Campo, R.; Brullet, E.; et al. Occult H. pylori infection partially explains ‘false-positive’ results of 13 C-urea breath test. United Eur. Gastroenterol. J. 2015, 3, 437–442. [Google Scholar] [CrossRef] [PubMed]
- Non, H. Pylori Bacteria with Urease Activity Identified in the Human Stomach. Nat. Clin. Pract. Gastroenterol. Hepatol. 2006, 3, 605. [Google Scholar] [CrossRef]
- Arama, S.S.; Tiliscan, C.; Negoita, C.; Croitoru, A.; Arama, V.; Mihai, C.M.; Pop, F.; Garg, A. Efficacy of 7-Day and 14-Day Triple Therapy Regimens for the Eradication of Helicobacter pylori: A Comparative Study in a Cohort of Romanian Patients. Gastroenterol. Res. Pract. 2016, 2016, 5061640. [Google Scholar] [CrossRef] [PubMed]
- Langdon, A.; Crook, N.; Dantas, G. The effects of antibiotics on the microbiome throughout development and alternative approaches for therapeutic modulation. Genome Med. 2016, 8, 39. [Google Scholar] [CrossRef] [PubMed]
- Dingsdag, S.A.; Hunter, N. Metronidazole: An update on metabolism, structure–cytotoxicity and resistance mechanisms. J. Antimicrob. Chemother. 2017, 73, 265–279. [Google Scholar] [CrossRef]
- Rizvanov, A.A.; Haertlé, T.; Bogomolnaya, L.; Abadi, A.T.B. Helicobacter pylori and Its Antibiotic Heteroresistance: A Neglected Issue in Published Guidelines. Front. Microbiol. 2019, 10, 1796. [Google Scholar] [CrossRef]
- Boyanova, L.; Hadzhiyski, P.; Gergova, R.; Markovska, R. Evolution of Helicobacter pylori Resistance to Antibiotics: A Topic of Increasing Concern. Antibiotics 2023, 12, 332. [Google Scholar] [CrossRef]
- Kaklikkaya, N.; Cubukcu, K.; Aydin, F.; Bakir, T.; Erkul, S.; Tosun, I.; Topbas, M.; Yazici, Y.; Buruk, C.K.; Erturk, M. Significance of cagA status and vacA subtypes of Helicobacter pylori in determining gastric histopathology: Virulence markers of H. pylori and histopathology. J. Gastroenterol. Hepatol. 2006, 21, 1042–1047. [Google Scholar] [CrossRef]
- Liu, X.; Nie, W.; Liang, J.; Li, Y. Interaction of Helicobacter Pylori with Other Microbiota Species in the Development of Gastric Cancer. Arch. Clin. Microbiol. 2017, 8, 67. [Google Scholar] [CrossRef]
- Vital, J.S.; Tanoeiro, L.; Lopes-Oliveira, R.; Vale, F.F. Biomarker Characterization and Prediction of Virulence and Antibiotic Resistance from Helicobacter pylori Next Generation Sequencing Data. Biomolecules 2022, 12, 691. [Google Scholar] [CrossRef]
- Nogueira, C.; Figueiredo, C.; Carneiro, F.; Gomes, A.T.; Barreira, R.; Figueira, P.; Salgado, C.; Belo, L.; Peixoto, A.; Bravo, J.C.; et al. Helicobacter pylori Genotypes May Determine Gastric Histopathology. Am. J. Pathol. 2001, 158, 647–654. [Google Scholar] [CrossRef] [PubMed]
- Roesler, B.M.; Rabelo-Gonçalves, E.M.; Zeitune, J.M. Virulence Factors of Helicobacter pylori: A Review. Clin. Med. Insights Gastroenterol. 2014, 7, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Demırtürk, L.; Özel, A.M.; Yazgan, Y.; Solmazgül, E.; Yildirim; Gültepe, M.; Gürbüz, A.K. CagA Status in Dyspeptic Patients with and without Peptic Ulcer Disease in Turkey: Association with Histopathologic Findings. Helicobacter 2001, 6, 163–168. [Google Scholar] [CrossRef]
- Norma, U.E.; Angelina, C.J.N.; Lilia, M.V.L.V.L.; Mayra, C.L.C.; Aurora, M.N.R.; Joseacute, A.H.H.N.; Juan, A.R.C.; Estela, R.B. Prevalence of Helicobacter pylori cagA and vacA genotypes in a population from Northeastern Mexico with chronic gastritis and intestinal metaplasia. Afr. J. Microbiol. Res. 2013, 7, 1409–1414. [Google Scholar] [CrossRef]
- Buffie, C.G.; Jarchum, I.; Equinda, M.; Lipuma, L.; Gobourne, A.; Viale, A.; Ubeda, C.; Xavier, J.; Pamer, E.G. Profound Alterations of Intestinal Microbiota following a Single Dose of Clindamycin Results in Sustained Susceptibility to Clostridium difficile-Induced Colitis. Infect. Immun. 2012, 80, 62–73. [Google Scholar] [CrossRef]
- Duynhoven, Y.T.V.; Jonge, R.D. Transmission of Helicobacter Pylori: A Role for Food? Bull. World Health Organ. 2001, 79, 455–460. [Google Scholar]
- Akamatsu, T.; Tabata, K.; Hironga, M.; Kawakami, H.; Yyeda, M. Transmission of Helicobacter pylori infection via flexible fiberoptic endoscopy. Am. J. Infect. Control. 1996, 24, 396–401. [Google Scholar] [CrossRef]
- Kim, D.-H.; Son, B.K.; Min, K.-W.; Han, S.K.; Na, J.U.; Choi, P.C.; Kim, H.-L.; Kwon, M.J.; Oh, Y.H.; Jung, W.Y.; et al. Chronic Gastritis Is Associated with a Decreased High-Density Lipid Level: Histological Features of Gastritis Based on the Updated Sydney System. J. Clin. Med. 2020, 9, 1856. [Google Scholar] [CrossRef]
- Kamada, N.; Chen, G.Y.; Inohara, N.; Núñez, G. Control of pathogens and pathobionts by the gut microbiota. Nat. Immunol. 2013, 14, 685–690. [Google Scholar] [CrossRef]
- Birg, A.; Ritz, N.L.; Lin, H.C. The Unknown Effect of Antibiotic-Induced Dysbiosis on the Gut Microbiota. Microbiome Metabolome Diagn. Ther. Other Strateg. Appl. 2019, 195–200. [Google Scholar] [CrossRef]
- Reed, T.; Lushington, G.H.; Xia, Y.; Hirakawa, H.; Travis, D.M.; Mure, M.; Scott, E.E.; Limburg, J. Crystal Structure of Histamine Dehydrogenase from Nocardioides simplex. J. Biol. Chem. 2010, 285, 25782–25791. [Google Scholar] [CrossRef]
- Hagiya, H.; Sugiyama, J.; Kuroe, Y.; Nojima, H.; Naito, H.; Hagioka, S.; Morimoto, N.; Murase, T. Delftia acidovorans bacteremia caused by bacterial translocation after organophosphorus poisoning in an immunocompetent adult patient. J. Infect. Chemother. 2013, 19, 338–341. [Google Scholar] [CrossRef]
- Ghattargi, V.C.; Nimonkar, Y.S.; Sape, K.; Prakash, O.; Suryavanshi, M.V.; Shouche, Y.S.; Meti, B.S.; Pawar, S.P. Functional and Comparative Genomics of Niche-Specific Adapted Actinomycetes Kocuria rhizophila Strain D2 Isolated from Healthy Human Gut. bioRxiv 2018, 400242. [Google Scholar] [CrossRef]
- Kuda, T. Quality Improvement and Fermentation Control in Fish Products. In Advances in Fermented Foods and Beverages: Improving Quality, Technologies and Health Benefits; Woodhead Publishing: Cambridge, UK, 2015; pp. 377–390. [Google Scholar] [CrossRef]
- Gomi, A.; Harima-Mizusawa, N.; Shibahara-Sone, H.; Kano, M.; Miyazaki, K.; Ishikawa, F. Effect of Bifidobacterium bifidum BF-1 on gastric protection and mucin production in an acute gastric injury rat model. J. Dairy Sci. 2013, 96, 832–837. [Google Scholar] [CrossRef] [PubMed]
- O’Callaghan, A.; van Sinderen, D. Bifidobacteria and Their Role as Members of the Human Gut Microbiota. Front. Microbiol. 2016, 7, 925. [Google Scholar] [CrossRef] [PubMed]
- Butta, H.; Sardana, R.; Vaishya, R.; Singh, K.N.; Mendiratta, L. Bifidobacterium: An Emerging Clinically Significant Metronidazole-resistant Anaerobe of Mixed Pyogenic Infections. Cureus 2017, 9, e1134. [Google Scholar] [CrossRef] [PubMed]
- Esaiassen, E.; Hjerde, E.; Cavanagh, J.P.; Simonsen, G.S.; Klingenberg, C. Bifidobacterium Bacteremia: Clinical Characteristics and a Genomic Approach To Assess Pathogenicity. J. Clin. Microbiol. 2017, 55, 2234–2248. [Google Scholar] [CrossRef]
- Segers, P.; Vancanneyt, M.; Pot, B.; Torck, U.; Hoste, B.; Dewettinck, D.; Falsen, E.; Kersters, K.; DE Vos, P. Classification of Pseudomonas diminuta Leifson and Hugh 1954 and Pseudomonas vesicularis Busing, Doll, and Freytag 1953 in Brevundimonas gen. nov. as Brevundimonas diminuta comb. nov. and Brevundimonas vesicularis comb. nov., Respectively. Int. J. Syst. Evol. Microbiol. 1994, 44, 499–510. [Google Scholar] [CrossRef]
- Schwartz, M.A.; Tabet, S.R.; Collier, A.C.; Wallis, C.K.; Carlson, L.C.; Nguyen, T.T.; Kattar, M.M.; Coyle, M.B. Central Venous Catheter–Related Bacteremia Due to Tsukamurella Species in the Immunocompromised Host: A Case Series and Review of the Literature. Clin. Infect. Dis. 2002, 35, e72–e77. [Google Scholar] [CrossRef]
- Lee, M.; Srinivasan, S.; Kim, M.K. New taxa in Alphaproteobacteria: Brevundimonas olei sp. nov., an esterase-producing bacterium. J. Microbiol. 2010, 48, 616–622. [Google Scholar] [CrossRef]
- Kandi, V.; Palange, P.; Vaish, R.; Bhatti, A.B.; Kale, V.; Kandi, M.R.; Bhoomagiri, M.R. Emerging Bacterial Infection: Identification and Clinical Significance of Kocuria Species. Cureus 2016, 8, e731. [Google Scholar] [CrossRef] [PubMed]
- Alkanany, F.N.; Gmais, S.A.; Maki, A.A.; Altaee, A.M. Estimation of Bacterial Biodegradability of PAH in Khor Al-Zubair Channel, Southern Iraq. Int. J. Mar. Sci. 2017, 7, 42. [Google Scholar] [CrossRef]
- Wisplinghoff, H. Pseudomonas spp., Acinetobacter spp. and Miscellaneous Gram-Negative Bacilli. In Infectious Diseases; Elsevier: Amsterdam, The Netherlands, 2017; pp. 1579–1599.e2. [Google Scholar] [CrossRef]
- Peduto, L.; Philippe, J.S.; Giulia, N. Chapter Three-Intestinal Stem Cells and Their Niche at Homeo-stasis and Under Stress. In Advances in Stem Cells and Their Niches; Bonnet, D., Ed.; Elsevier: Amsterdam, The Netherlands, 2018; Volume 2, pp. 77–97. [Google Scholar] [CrossRef]
- Ryan, M.P.; Pembroke, J.T. Brevundimonas spp.: Emerging global opportunistic pathogens. Virulence 2018, 9, 480–493. [Google Scholar] [CrossRef] [PubMed]
- Palleja, A.; Mikkelsen, K.H.; Forslund, S.K.; Kashani, A.; Allin, K.H.; Nielsen, T.; Hansen, T.H.; Liang, S.; Feng, Q.; Zhang, C.; et al. Recovery of gut microbiota of healthy adults following antibiotic exposure. Nat. Microbiol. 2018, 3, 1255–1265. [Google Scholar] [CrossRef] [PubMed]
- Pettigrew, M.M.; Gent, J.F.; Kong, Y.; Halpin, A.L.; Pineles, L.; Harris, A.D.; Johnson, J.K. Gastrointestinal Microbiota Disruption and Risk of Colonization with Carbapenem-resistant Pseudomonas aeruginosa in Intensive Care Unit Patients. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2018, 69, 604–613. [Google Scholar] [CrossRef]
- Cogen, A.L.; Nizet, V.; Gallo, R.L. Skin microbiota: A source of disease or defence? Br. J. Dermatol. 2008, 158, 442–455. [Google Scholar] [CrossRef]
- Wagner, J.; Short, K.; Catto-Smith, A.G.; Cameron, D.J.S.; Bishop, R.F.; Kirkwood, C.D. Identification and Characterisation of Pseudomonas 16S Ribosomal DNA from Ileal Biopsies of Children with Crohn’s Disease. PLoS ONE 2008, 3, e3578. [Google Scholar] [CrossRef]
- Gao, Y.-D.; Zhao, Y.; Huang, J. Metabolic Modeling of Common Escherichia coli Strains in Human Gut Microbiome. BioMed Res. Int. 2014, 2014, 694967. [Google Scholar] [CrossRef]
- Conway, T.; Cohen, P.S. Commensal and Pathogenic Escherichia coli Metabolism in the Gut. Microbiol. Spectr. 2015, 3, 343–362. [Google Scholar] [CrossRef]
- Sonnenborn, U. Escherichia coli strain Nissle 1917—From bench to bedside and back: History of a special Escherichia coli strain with probiotic properties. FEMS Microbiol. Lett. 2016, 363, fnw212. [Google Scholar] [CrossRef]
- Fang, K.; Jin, X.; Hong, S.H. Probiotic Escherichia coli inhibits biofilm formation of pathogenic E. coli via extracellular activity of DegP. Sci. Rep. 2018, 8, 4939. [Google Scholar] [CrossRef] [PubMed]
- Dutkiewicz, J.; Mackiewicz, B.; Lemieszek, M.K.; Golec, M.; Milanowski, J. Pantoea agglomerans: A mysterious bacterium of evil and good. Part IV. Beneficial effects. Ann. Agric. Environ. Med. AAEM 2016, 23, 206–222. [Google Scholar] [CrossRef] [PubMed]
- Büyükcam, A.; Tuncer, Ö.; Gür, D.; Sancak, B.; Ceyhan, M.; Cengiz, A.B.; Kara, A. Clinical and microbiological characteristics of Pantoea agglomerans infection in children. J. Infect. Public Health 2018, 11, 304–309. [Google Scholar] [CrossRef] [PubMed]
- Delgado, S.; Cabrera-Rubio, R.; Mira, A.; Suárez, A.; Mayo, B. Microbiological Survey of the Human Gastric Ecosystem Using Culturing and Pyrosequencing Methods. Microb. Ecol. 2013, 65, 763–772. [Google Scholar] [CrossRef]
- Kovaleva, J.; Degener, J.E.; van der Mei, H.C. Methylobacterium and Its Role in Health Care-Associated Infection. J. Clin. Microbiol. 2014, 52, 1317–1321. [Google Scholar] [CrossRef]
- Mukherjee, S.; Ramesh, A. Bacteriocin-producing strains of Lactobacillus plantarum inhibit adhesion of Staphylococcus aureus to extracellular matrix: Quantitative insight and implications in antibacterial therapy. J. Med. Microbiol. 2015, 64, 1514–1526. [Google Scholar] [CrossRef]
- Mohajeri, M.H.; Brummer, R.J.M.; Rastall, R.A.; Weersma, R.K.; Harmsen, H.J.M.; Faas, M.; Eggersdorfer, M. The role of the microbiome for human health: From basic science to clinical applications. Eur. J. Nutr. 2018, 57 (Suppl. 1), 1–14. [Google Scholar] [CrossRef]
- Liu, J.; Xue, Y.; Zhou, L. Detection of gastritis-associated pathogens by culturing of gastric juice and mucosa. Int. J. Clin. Exp. Pathol. 2018, 11, 2214–2220. [Google Scholar]
- Barbosa, A.J.A.; Boldt, M.S.; Rodrigues, C.B.; Silva, C.S.C.; Ferreira, H.M.; Pereira, R.D. Histopathological features of mucosa atrophy in atrophic body gastritis. J. Bras. De Patol. E Med. Lab. 2016, 52, 50–54. [Google Scholar] [CrossRef]
- Jonaitis, P.; Kupcinskas, L.; Kupcinskas, J. Molecular Alterations in Gastric Intestinal Metaplasia. Int. J. Mol. Sci. 2021, 22, 5758. [Google Scholar] [CrossRef]
- Schubert, J.P.; Rayner, C.K.; Costello, S.P.; Roberts-Thomson, I.C.; Forster, S.C.; Bryant, R.V. Helicobacter pylori: Have potential benefits been overlooked? JGH Open 2022, 6, 735–737. [Google Scholar] [CrossRef] [PubMed]
- Nordenstedt, H.; Graham, D.Y.; Kramer, J.R.; Rugge, M.; Verstovsek, G.; Fitzgerald, S.; Alsarraj, A.; Shaib, Y.; Velez, M.E.; Abraham, N.; et al. Helicobacter pylori -Negative Gastritis: Prevalence and Risk Factors. Am. J. Gastroenterol. 2013, 108, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Miftahussurur, M.; Waskito, L.A.; Syam, A.F.; Nusi, I.A.; Wibawa, I.D.N.; Rezkitha, Y.A.A.; Siregar, G.; Yulizal, O.; Akil, F.; Uwan, W.B.; et al. Analysis of risks of gastric cancer by gastric mucosa among Indonesian ethnic groups. PLoS ONE 2019, 14, e0216670. [Google Scholar] [CrossRef] [PubMed]
- Waskito, L.A.; Rezkitha, Y.A.A.; Vilaichone, R.-K.; Sugihartono, T.; Mustika, S.; Wibawa, I.D.N.; Yamaoka, Y.; Miftahussurur, M. The role of non-Helicobacter pylori bacteria in the pathogenesis of gastroduodenal diseases. Gut Pathog. 2022, 14, 19. [Google Scholar] [CrossRef]
- Miftahussurur, M.; Waskito, L.A.; El-Serag, H.B.; Ajami, N.J.; Nusi, I.A.; Syam, A.F.; Matsumoto, T.; Rezkitha, Y.A.A.; Doohan, D.; Fauzia, K.A.; et al. Gastric microbiota and Helicobacter pylori in Indonesian population. Helicobacter 2020, 25, e12695. [Google Scholar] [CrossRef]
- Gantuya, B.; El-Serag, H.B.; Matsumoto, T.; Ajami, N.J.; Oyuntsetseg, K.; Azzaya, D.; Uchida, T.; Yamaoka, Y. Gastric Microbiota in Helicobacter pylori-Negative and -Positive Gastritis Among High Incidence of Gastric Cancer Area. Cancers 2019, 11, 504. [Google Scholar] [CrossRef]
- Zhang, X.; Li, C.; Cao, W.; Zhang, Z. Alterations of Gastric Microbiota in Gastric Cancer and Precancerous Stages. Front. Cell. Infect. Microbiol. 2021, 11, 559148. [Google Scholar] [CrossRef]
- Kim, H.-N.; Kim, M.-J.; Jacobs, J.P.; Yang, H.-J. Altered Gastric Microbiota and Inflammatory Cytokine Responses in Patients with Helicobacter pylori-Negative Gastric Cancer. Nutrients 2022, 14, 4981. [Google Scholar] [CrossRef]

