1. Introduction
The study of the microbiota in liver diseases has gained significant interest due to the emerging understanding of the intricate relationship between gut microbial communities and liver health. Studies have investigated the significant function of the gut–liver axis as a feedback loop in the gastrointestinal system. It is thought that alterations in the gut microbiota are closely associated with an increase in intestinal permeability. This alteration can lead to the release of endotoxins, triggering inflammatory responses and affecting bile acid metabolism [1]. Accumulating evidence suggests that the gut–liver axis plays a pivotal role in various liver conditions, from Alcoholic Liver Disease (ALD) and Non-alcoholic Fatty Liver Disease (NAFLD) to Hepatocellular Carcinoma (HCC) [2,3]. Dysbiosis, a term used to describe the imbalance of the gut microbiota, has been linked to inflammation, metabolic dysfunction, and the progression of liver diseases [4]. Investigating the composition and function of the microbiota in these contexts holds promise for identifying novel biomarkers, therapeutic targets, and interventions that could revolutionize the management and prevention of liver diseases.
One of the key members of the microbiota in liver diseases is the genus Bifidobacterium. Bifidobacterium is an essential genus of bacteria found in the gastrointestinal tract. Its main protective mechanisms include improved adherence to the intestinal barrier, increased production of short-chain fatty acid (SCFA) and pH reduction, the release of bifidocins (peptides that inhibit the growth of other pathogenic bacteria), and positive immunomodulatory effects [5]. Nevertheless, the levels of Bifidobacterium are often reduced in liver injuries, which may be directly linked to the pathophysiology of the disease. Consequently, recent studies have focused on how probiotics can be used to supplement this deficiency and potentially treat liver diseases more effectively [6].
Next-generation sequencing (NGS), also known as high-throughput DNA sequencing, has become the preferred method for analyzing microbiotas in various human pathologies, including liver diseases. The most common approach to identifying bacteria based on these modern technologies is through amplicon sequencing or shotgun metagenomics. Amplicon sequencing involves PCR amplification and sequencing of the 16S ribosomal RNA (16S rRNA) gene [7]. This technique is relatively cost-effective and provides a targeted analysis of the microbial taxa in each sample, providing valuable insights into the composition and diversity of the microbiota associated with different clinical conditions [8]. However, shotgun metagenomics is an alternative approach in which the entire microbial DNA is sequenced without any prior amplification or targeting. This method provides a more comprehensive view of the microbial community, including bacteria, viruses, fungi, and other microorganisms [9]. Shotgun metagenomics allows for the identification of novel pathogens and the exploration of functional potentials within the microbiota [10]. However, it can be more expensive and computationally intensive compared to amplicon sequencing.
For nearly a decade, NGS technologies have been used to study the bacterial communities in liver diseases [11]. These studies have employed different sampling techniques, sequencing technologies, and statistical methods. The diversity of these factors makes it challenging to obtain a comprehensive overview of the field and identify the main trends in the microbial composition associated with each stage of the disease, especially when it comes to scientific but relevant taxa such as the genus Bifidobacterium.
In this systematic review, we conducted a comprehensive and critical review of research that has explored the relationship between the Bifidobacterium genus and liver diseases, from NAFLD and ALD to HCC. We specifically focused on studies that used next-generation sequencing (NGS) technologies to investigate this association, providing, for the first time, an in-depth analysis of the methods in liver diseases and the identification of this particular genus. Our goal was to analyze and combine the results of studies that explore the potential impact of Bifidobacterium on liver damage and its possible correlation with disease advancement. We conducted a thorough review of existing literature to achieve this objective. By studying how Bifidobacterium exerts these effects, we can gain a better understanding of the underlying mechanisms of liver injuries and potentially develop new treatments that target these mechanisms.
2. Materials and Methods
2.1. Search Strategy and Selection Criteria
A systematic literature search was conducted using the PubMed, Embase, and Web of Science databases. The investigation was limited to articles published in English from January 2013 to June 2023. The search terms included “Bifidobacterium”, “NAFLD”, “NASH”, “ALD”, “cirrhosis”, and “HCC”, along with their medical subject headings (MeSH in PubMed and Emtree in Embase). The search strategy was adapted and standardized across all databases with careful application of logical connectors. The contains a detailed search strategy for each database. Studies that did not meet these criteria were excluded, excluding reviews, case reports, editorials, book chapters, conference abstracts, and notes from the outset. In particular, we focused on studies that used next-generation sequencing (NGS) technologies to investigate the potential impact of Bifidobacterium on liver damage. Six hundred forty-eight initial studies were identified through a customized strategy for systematic database search, as shown in Figure 1.
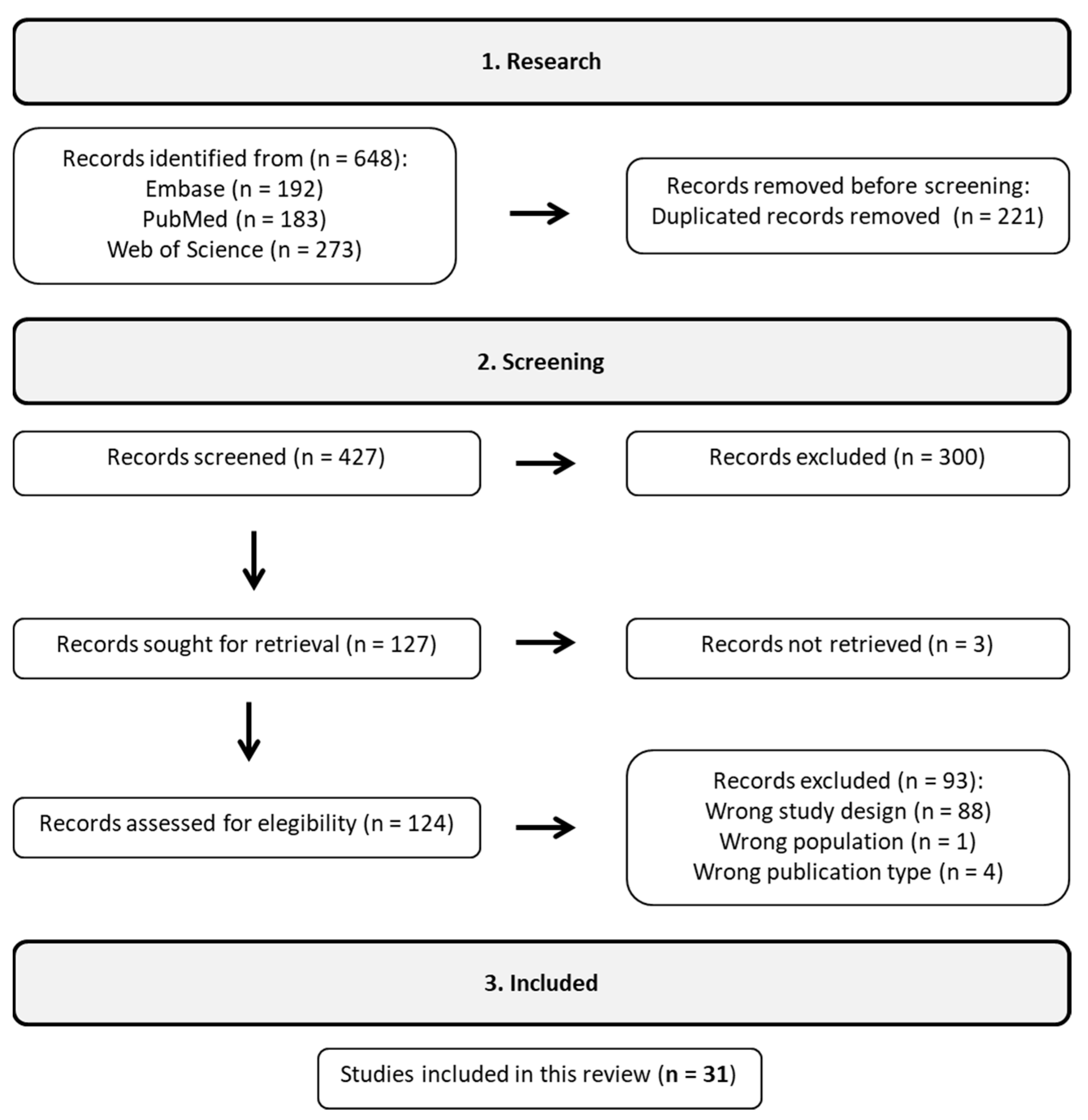
Figure 1. According to the workflow, 31 studies were included out of the initial 648 following a rigorous filtering process.
The Rayyan web platform was used to identify and remove duplicate reports [12]. Initially, article relevance was evaluated based on their titles and abstracts, and a comprehensive content analysis was conducted when necessary. The selected papers then underwent a thorough assessment, with relevant data extracted for research analysis. After removing duplicates and studies that did not fit the scope of this review, a total of 31 studies were selected to form the representative sample. Subsequently, these studies were organized according to their respective categories: observational and observational with treatment. The Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) criteria were carefully adhered to in this review, as shown in [13].
2.2. Data Extraction and Analysis
The data from the selected articles were organized into a dataset containing the following variables: authors’ names, publication year, country, disease, population (human or animal), type of study (epidemiological classification), sample type, next-generation sequencing technology, sequencing type, sample size (N), groups, statistical methodology, database, and clinical outcomes. These data are available in .
Risk-of-bias analysis was conducted using validated approaches from the Joanna Briggs Institute (JBI) to assess the quality of studies [14]. JBI tools provide robust criteria and standards to guide study quality.
3. Results
3.1. Selected Studies
presents the results of 17 studies that evaluated the levels of Bifidobacterium in both control and disease groups, encompassing human and animal samples. These findings were developed in various countries, with China leading the number of publications.
In , although the studies presented disease groups separately, the Bifidobacterium levels for those groups were not measured. Only the disease group with treatment had available abundance values for Bifidobacterium.
In general, the 31 studies used two types of sequencing: 16S rRNA amplicon sequencing and shotgun metagenomics (Figure 2A). In 16S rRNA sequencing, the most prevalent region was V3–V4, followed by V4. Regions V1–V3 and V4–V5 appeared less frequently. There was considerable variability in the databases used for taxonomic classification (Figure 2A). The most commonly used databases for 16S rRNA classification were SILVA and Greengenes, while MetaPhIAn was used for shotgun metagenomic analysis. It is noteworthy that several studies did not report the databases used. Concerning the statistical analysis used to identify differentially abundant genera between control and disease groups, Linear Discriminant Analysis of Effect Size (LEfSe) and Mann–Whitney were the most employed methods (Figure 2A). However, many studies also resorted to simple statistical tests, such as Mann–Whitney, Wilcoxon, Kruskal–Wallis, and Student’s t-test. Once again, many studies did not report the statistical methods used for abundance differentiation. These findings highlight the diversity of methodological approaches used in Bifidobacterium studies and the need for greater transparency in disclosing the methods employed to strengthen the reliability and comparability of the results obtained.
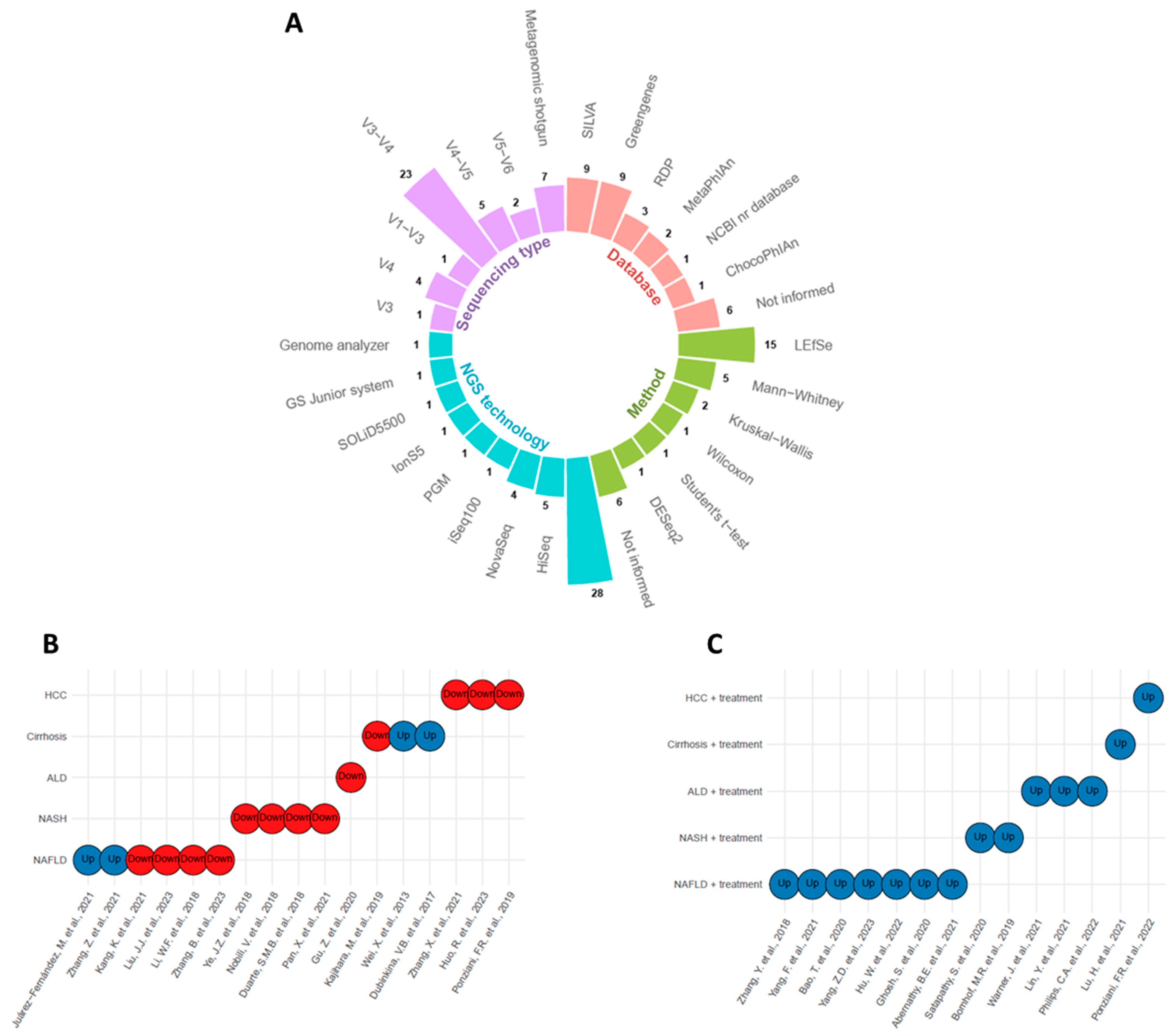
Figure 2. Overview of the methods used to detect and quantify Bifidobacterium genus in liver lesions and their upregulation or downregulation status. (A) Circular barplot with sequencing type, database, method, and next-generation sequencing (NGS) technology information. (B) Bifidobacterium levels in untreated disease groups [4,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30], see . (C) Bifidobacterium levels in treated disease groups [31,32,33,34,35,36,37,38,39,40,41,42,43,44], see . Blue circles denote upregulation, and red circles indicate downregulation.
The levels of Bifidobacterium in liver diseases were assessed in several observational studies. Out of 17 studies, only four reported elevated levels of Bifidobacterium in the disease group compared to the control group (Figure 2B). Two of these studies involved NAFLD and the other two involved cirrhosis. In contrast, all other studies reported reduced levels of Bifidobacterium in the disease groups compared to control.
In addition, 14 studies observed Bifidobacterium levels in the disease groups after receiving some form of treatment, such as a drug, molecule, or probiotic that did not contain Bifidobacterium in its composition (Figure 2C). All these studies demonstrated an increase in Bifidobacterium levels after treatment.
3.2. Risk-of-Bias and Quality Assessment
Out of the 31 studies reviewed, 17 followed a clinical model and were categorized by their epidemiological approaches: case-control, cross-sectional, and cohort. The case-control and cross-sectional studies had a high risk of bias levels, while the cohort studies exhibited a low risk of bias. All information regarding bias risk is displayed in Figure 3.
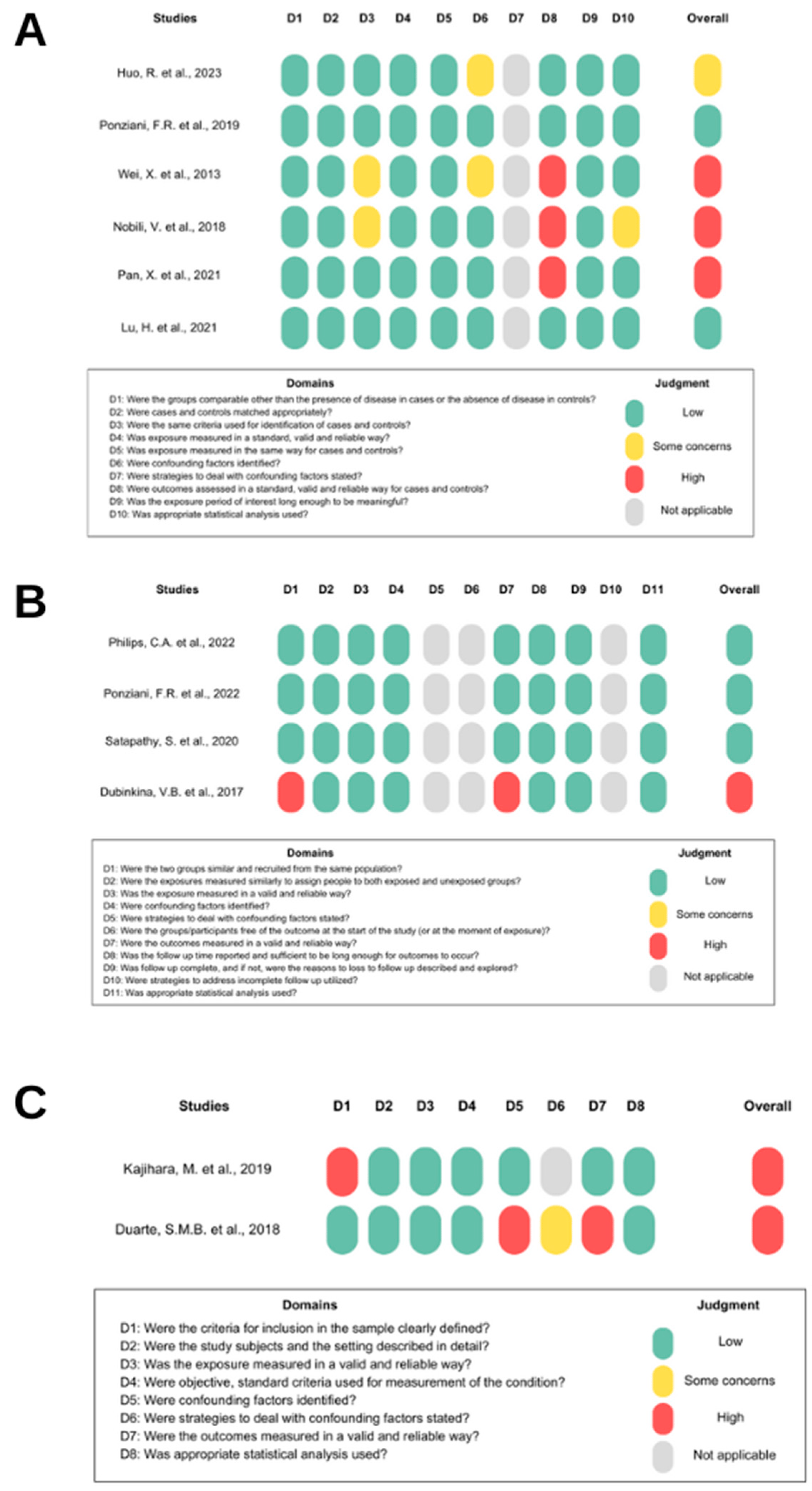
Figure 3. Risk of bias. (A) shows the risk of bias in case-control studies [4,19,28,30,34,35], (B) in cohort studies [20,33,34,43], and (C) in cross-sectional studies [18,29]. Domain “D” represents the items in the JBI checklist. “Overall” represents the study’s risk of bias based on judgment criteria.
4. Discussion
This review highlights the strong relationship between the Bifidobacterium genus and different hepatic lesions (including ALD, NAFLD, NASH, cirrhosis, and HCC). Remarkably, most studies have revealed significantly reduced levels of Bifidobacterium in these hepatic conditions compared to control groups. Additionally, all studies that employed treatments for these diseases observed restoring Bifidobacterium levels. These results strongly support the hypothesis that this bacterial genus plays a pivotal role in the health status of humans as a member of the beneficial microbiota, and major changes in its abundance significantly influence both the pathophysiology of hepatic diseases and their therapeutic approach. Furthermore, we thoroughly evaluated the NGS methodologies and associated statistical analyses employed in each study. We specifically examined the methods used to determine the differential abundance of Bifidobacterium, highlighting concerns such as potential biases and the omission of relevant information in certain studies.
The discussion section was organized to comprehensively understand the relationship between Bifidobacterium and the pathophysiology of each liver injury. Initially, the section presented the role of Bifidobacterium in maintaining the balance of the intestinal microbiota and its contribution to a healthy phenotype. Subsequently, the relationship between Bifidobacterium and different liver conditions was addressed, covering the progression and stages of liver diseases in Section 4.1, the Bifidobacterium genus in Section 4.2, ALD in Section 4.3, NAFLD in Section 4.4, NASH in Section 4.5, cirrhosis in Section 4.6, and HCC in Section 4.7. Each section explored the underlying mechanisms of action and the potential therapeutic effects of Bifidobacterium for each liver condition. This approach aims to provide a comprehensive understanding of the influence of Bifidobacterium on each liver injury and its potential clinical implications.
4.1. The Progression and Stages of Liver Diseases
The liver plays a crucial role in digestion and metabolism, making it vulnerable to the effects of diet and alcohol consumption. Although the physiological processes share common mechanisms, it is possible to identify a non-alcoholic pathway, more closely related to the excessive accumulation of fat, and an alcoholic pathway, triggered by the toxicity of excessive alcohol consumption (Figure 4). The non-alcoholic pathway progresses through the stages of NAFLD and NASH, ultimately leading to extensive fibrous tissue and the characteristic inflammatory process of a cirrhotic liver [45].
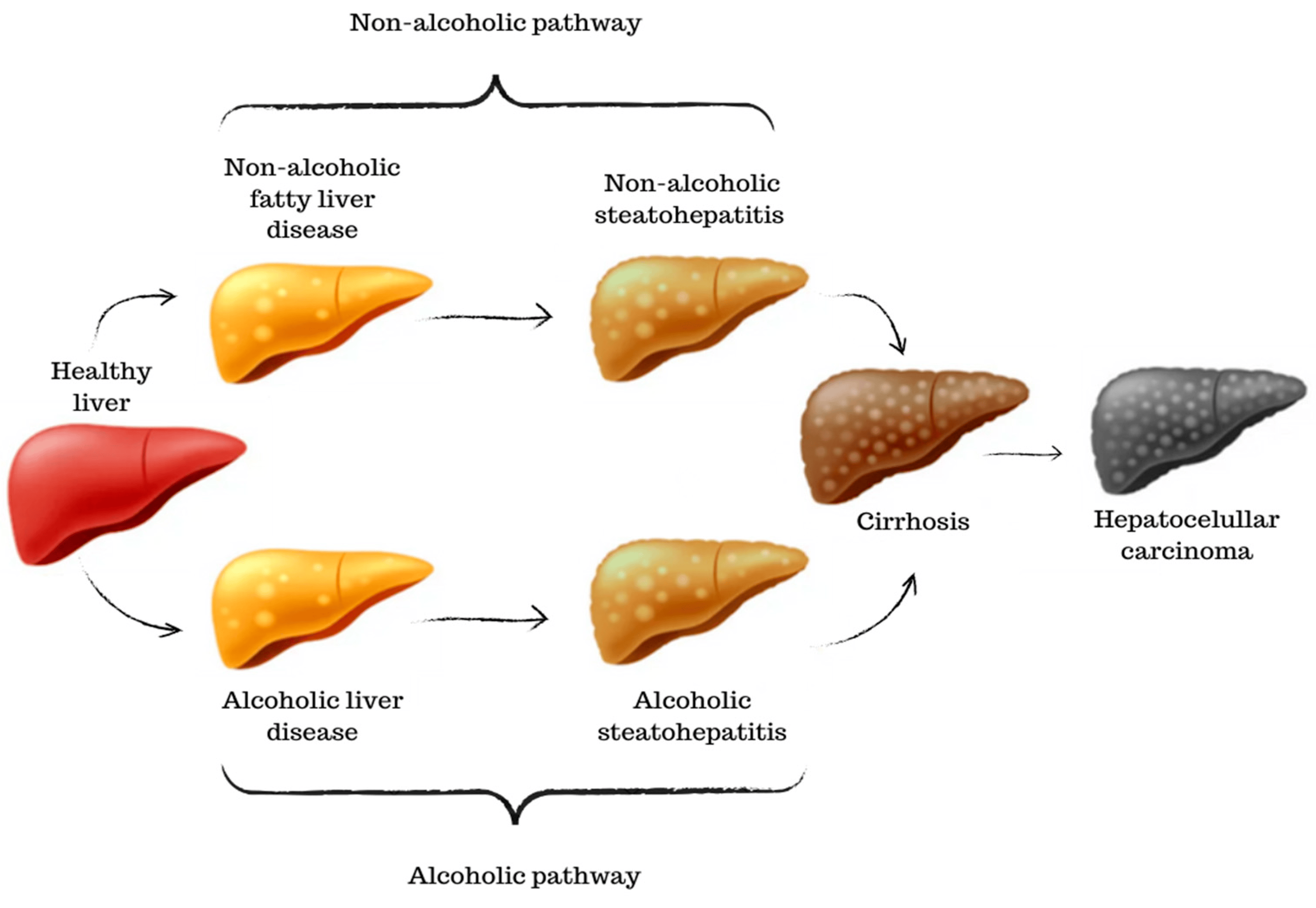
Figure 4. A graphical scheme of the progression and stages of liver diseases. Starting from a healthy liver until the development of hepatocellular carcinoma.
Similar to NAFLD, the alcoholic pathway begins with the accumulation of fat in liver cells due to excessive alcohol consumption. This condition is reversible if alcohol consumption is stopped. Prolonged and heavy alcohol consumption can lead to inflammation and liver cell damage. This stage is known as alcoholic steatohepatitis. Some individuals with alcoholic steatohepatitis can experience rapid disease progression. Advanced fibrosis can progress to cirrhosis in ALD as well [46]. In both pathways, the severity of cirrhosis can lead to HCC, the most advanced stage of liver cancer, as well as a loss of liver function and, often, death [47].
4.2. The Bifidobacterium Genus
In 1900, Henri Tissier discovered a rod-shaped bacterium initially thought to belong to the Lactobacteriaceae family. This bacterium was gram-positive, catalase-negative, anaerobic, non-spore-forming, non-gas-producing, and non-motile. After new research emerged, it was determined that Bifidobacterium is a genus that pertains to the Actinobacteria phylum, thereby distinguishing bifidobacteria from lactobacilli [48]. According to the List of Prokaryotic Names with Standing in Nomenclature (LPSN), about 90 species have been identified until now, with some of the most-studied and well-known being B. adolescentis, B. infantis, B. bifidum, B. breve, and B. longum [49].
Bifidobacterium constitutes a relevant part of the human body’s microbiota and is present in various regions of the organism, such as the oral cavity, gastrointestinal tract, vagina, and cervix [50]. At the time of birth, due to fluid exchange between mother and fetus, the levels of Bifidobacterium are higher. As the child develops and transitions to adulthood, there is a separation from maternal feeding and consequent ingestion of nutritionally poor foods, resulting in a drastic reduction in Bifidobacterium levels [51].
The presence of the Bifidobacterium genus in the composition of the human gastrointestinal microbiota has been associated with numerous health benefits. For instance, in the human diet, undigested dietary fibers are fermented by Bifidobacterium. In this process, SCFAs are produced, which, in turn, are associated with intestinal cell health and immune system regulation [52]. Moreover, the mere presence of Bifidobacterium helps maintain the microbiota balance as they compete for space and nutrients, limiting the growth of potentially harmful bacteria. Consequently, this leads to positive immune system modulation, intestinal barrier strengthening, and nutrient absorption improvement [53].
Additionally, studies have shown that levels of Bifidobacterium are reduced in various conditions such as diabetes and major depression [54,55]. Some analyses explore the potential of Bifidobacterium levels in aiding disease diagnosis and treatment [56].
4.3. Bifidobacterium in ALD
ALD is the result of chronic and excessive alcohol consumption. Although there is no well-defined minimum threshold, it is known that ingesting around 40 g of pure alcohol per day over many years significantly increases the risk of developing ALD [3]. ALD is a condition that varies in severity, ranging from early stages of hepatic steatosis to more severe complications, such as cirrhosis and HCC [57].
Changes in gut microbiota associated with ALD include increased gram-positive bacteria, such as the Enterobacteriaceae family, linked to a pro-inflammatory state. However, bacteria from the Clostridia class, which produce SCFAs like butyrate, are less abundant [20].
This review included only one study according to the established criteria. As expected, Bifidobacterium levels were reduced in the disease group, which correlated with a pronounced pro-inflammatory profile [15]. The decrease in Bifidobacterium in ALD is caused by alcohol toxicity, which disrupts gut mucosa and microbiota [58].
The hypothesis that reduced levels of Bifidobacterium in ALD are related to metabolic and microbiota imbalances gains more strength when observing the results of studies using treatments for the disease. In animal models, it was observed that the diseased group receiving human beta-defensin 2 (hBD-2) showed an increase in Bifidobacterium abundance, which may be associated with less hepatic fat accumulation, reduced hepatocellular injury, and inflammation [31]. In another animal study, treatment with auricularia auricula melanin (AMM) also elevated Bifidobacterium levels in the diseased group, along with reducing harmful bacteria presence and improving intestinal mucosal health [32]. In a clinical trial, ALD patients who received fecal microbiota transplantation also experienced increased levels of Bifidobacterium and improvements in digestive and immune health due to microbial modulation [33].
In conclusion, chronic alcohol consumption disrupts the gut microbiota, reducing the abundance of the beneficial genus Bifidobacterium and increasing pro-inflammatory bacteria. Studies show that treatments promoting Bifidobacterium increase can improve liver health and reduce inflammation in ALD.
4.4. Bifidobacterium in NAFLD
NAFLD is a hepatometabolic disease characterized by at least 5% steatosis in hepatocytes caused by abnormal fat accumulation. However, in the development of NAFLD, the fatty accumulation is not caused by excessive alcohol consumption or other autoimmune metabolic disorders, although the presence of comorbidities such as Type 2 diabetes and hypertension is familiar [59]. The presence of fat and the resulting metabolic alterations directly affect the loss of diversity and modification of the gastrointestinal microbiota composition [60].
This microbiota imbalance is related to the dysregulation in the production of SCFA and the inhibition of fasting-induced adiposity factor (FIAF) secretion. These changes lead to a biochemical cascade that induces fat accumulation in adipocytes and hepatocytes through lipogenesis. Additionally, dysbiosis is involved in increased lipopolysaccharide (LPS) production, which stimulates a pro-inflammatory response through Toll-like receptor 4 (TLR4) activation and promotes Kupffer cell activation. Interestingly, although NAFLD is not caused by excessive alcohol consumption, the bacteria, in the context of dysbiosis, can produce ethanol endogenously. This phenomenon increases intestinal permeability, toxins, and inflammation while reducing nutrient absorption [61].
Despite most studies reviewed in this article corroborating with the literature and pointing to a reduction in Bifidobacterium levels in NAFLD patients, two exceptional studies reported an increase in the levels of this bacterium. It is important to note that, in such exceptional cases, the increased levels of Bifidobacterium were not associated with harmful effects. This finding challenges the notion that any deviation in Bifidobacterium levels is detrimental, indicating a need for further research to comprehend the adaptive mechanisms of the microbiota in the context of NAFLD.
The discrepancies observed among studies regarding the levels of Bifidobacterium in NAFLD patients may be attributed to different factors. Firstly, the composition of the gut microbiota is highly individualized and can vary significantly between individuals [62]. This natural variation in gut microbiota composition can contribute to differences in Bifidobacterium levels observed among different study populations. Second, NAFLD itself is a complex condition with various underlying causes and disease progression patterns. Factors such as diet, lifestyle, genetics, and co-existing health conditions can influence the composition of the gut microbiota and its relationship with NAFLD [63]. The phenomenon of the adaptive response of the microbiota demonstrates its dynamic capacity to adapt to the environment and the inflammatory mechanisms of the human body [64]. Thus, variations in these factors among study populations may also contribute to the observed discrepancies. In addition, differences in the timing of sample collection and the duration of the study period may also contribute to the significant differences in results [65]. Lastly, the methods used to analyze and measure Bifidobacterium levels can vary between studies. Differences in sequencing techniques and data analysis can all influence the accuracy and comparability of the results. Therefore, inconsistencies in the methodologies employed by different research groups can contribute to the discrepancies observed.
Studies observing groups with NAFLD who received treatment have consistently shown increased Bifidobacterium levels. This supports the hypothesis that the disease’s development is linked to an imbalance in gut bacteria, specifically a decrease in Bifidobacterium. To achieve the best possible clinical results, it is essential to concentrate on restoring the levels of Bifidobacterium. Inulin, for example, has been a potential treatment for NAFLD, which has gained prominence in studies. It is a critical dietary fiber in the production of SCFA, stimulating the growth of Bifidobacterium. Consequently, there is an improvement in intestinal barrier health and the immune system [66].
In summary, NAFLD is linked to an imbalance in gut microbiota, specifically a decrease in Bifidobacterium levels and altered SCFA production. A potential approach to treating NAFLD is restoring Bifidobacterium levels through interventions like inulin. Studies on animals and humans have shown that certain Bifidobacterium species have inhibitory effects on NAFLD, indicating that gut microbiota is a promising target for therapy.
4.5. Bifidobacterium in NASH
NASH is a condition that is part of the NAFLD spectrum but is considered more severe due to the progression of inflammation and the development of liver lesions [67]. NAFLD is characterized by the accumulation of fat in the liver, which can evolve into NASH. In the latter, inflammation levels are typically higher, and intestinal permeability becomes more impaired, leading to a more significant imbalance in the microbiota [59].
Similar to NAFLD, in NASH, there is an increase in the genera Provatella and Escherichia, which are gram-negative bacteria associated, for example, with increased ethanol production [68]. However, the genus Bifidobacterium shows a decrease in abundance as the disease progresses [2]. All studies included in this review that evaluated Bifidobacterium levels in NASH confirmed these findings. This reduction in Bifidobacterium can exacerbate the microbiota imbalance, worsening intestinal permeability and directly impacting liver lipid accumulation, insulin resistance, and fibrosis development [59].
Observational studies on treatments included in this review support the idea that reducing Bifidobacterium levels harms the patient’s biochemical and clinical status. For example, after liver transplantation in NASH patients, an increase in Bifidobacterium levels was observed, possibly correlated with reduced hepatic fat [43]. Patients who received oligofructose also had their Bifidobacterium abundance restored, which was accompanied by reduced inflammation and fibrosis [44].
These findings emphasize the importance of the intestinal microbiota in NASH progression and suggest that strategies to preserve or restore Bifidobacterium levels may be beneficial in treating this liver condition.
4.6. Bifidobacterium in Cirrhosis
Cirrhosis is a clinical condition characterized by fibrosis resulting from recurrent wound-healing events in the hepatic tissue. These wounds are, in turn, a consequence of excessive fat accumulation and chronic alcohol exposure [69]. This context also promotes increased vascularization in the liver and portal hypertension [70]. Not surprisingly, liver transplantation remains the best curative option, although some drugs are already under study and show therapeutic potential [71].
Due to these pathophysiological mechanisms, cirrhosis is a disease intimately affected by the dysregulation of the gut microbiota. The simultaneous increase in Enterobacteriaceae, Enterococcaceae, and Streptococcaceae families, as well as the decrease in Lachnospiraceae and Ruminococcaceae families, strongly suggests elevated toxin production, decreased conversion of primary bile acids into secondary bile acids, and reduced production of SCFA [72].
In this review, the included observational studies did not provide conclusive results regarding the levels of Bifidobacterium in cirrhotic patients. One study reported a lower abundance of Bifidobacterium in patients, related to worsened inflammatory profile and liver damage [18]. On the other hand, two other studies found higher levels of Bifidobacterium in disease groups compared to control groups. Although it was a different result than expected, the researchers discuss that similar results have been found in other diseases, mainly cardiac and renal diseases. However, there is no correlation between a higher presence of Bifidobacterium and worse clinical outcomes in cirrhotic patients [19,20].
Although there is no clear definition of whether Bifidobacterium levels are reduced or increased in cirrhotic patients, treatment for the disease can elevate these levels. Furthermore, it appears that the cirrhotic patient’s health improves with increasing levels of Bifidobacterium. A recent study delved into the therapeutic effects of lactitol. The patients who were administered this treatment showed an abundance of Bifidobacterium and experienced improved health. The treatment also reduced the biosynthesis of endotoxin and inflammatory components [35].
Therefore, while studies confirmed that the intestinal microbiota affects cirrhosis, it is not established how Bifidobacterium changes in cirrhotic patients. More research is needed to better understand its role and therapeutic potential.
4.7. Bifidobacterium in HCC
HCC is the most common form of liver cancer in adults and is one of the leading causes of global mortality. In recent years, the predominant cause of HCC development has shifted from primarily being related to viral hepatitis to being associated with increased alcohol consumption and liver fat accumulation. Both patients with ALD and those with cirrhosis have a high risk of progression to HCC [73].
Excess alcohol and liver fat accumulation cause metabolic alterations that, over time, lead to crucial genomic changes in hepatocytes. The significant alterations involve genetic mutations, such as in the Telomerase Reverse Transcriptase (TERT) promoter and Tumor Protein 53 (TP53) gene, epigenetic modifications, and changes in growth factors, such as the Wnt/β-catenin and tyrosine kinase pathways [74].
This complex pathogenesis, with multiple interconnected mechanisms, also significantly affects the microbiota in HCC. Various imbalances are found, including the enrichment of bacteria such as Veillonella, Streptococcus, Clostridium, and Provotella, as well as the reduction of beneficial bacteria such as Eubacterium, Faecalibacterium, and Bifidobacterium [75].
Results from studies consistently demonstrated that both in animal models and clinical trials, Bifidobacterium levels are reduced in HCC [3,47,48]. An imbalanced microbiota, especially with lower levels of Bifidobacterium, can trigger a series of events, such as increased pathogenic bacteria, higher production of toxic and pro-carcinogenic substances, chronic inflammation, increased oxidative stress, DNA damage, progression of liver fibrosis, and cell death.
However, treatments with Tremelimumab and Durvalumab, two immunotherapeutic drugs, have shown clinical remission in HCC patients, and this effect may be related to the increase in Bifidobacterium levels observed after treatment. Beneficial bacteria such as Bifidobacterium and Akkermansia may enhance treatment response, mainly due to their anti-inflammatory activity and production of SCFA [34].
These findings highlight the importance of intestinal microbiota in HCC pathogenesis and treatment. They may provide insights for developing new therapeutic approaches based on the manipulation of Bifidobacterium to improve clinical outcomes for these patients.
4.8. Risk-of-Bias and Quality Assessment
Only three studies identified confounding factors. Huo, R. et al. reported that patients with advanced HCC received various treatments [4]. Wei, X. et al. also emphasized that using medications, diverse diets, and comorbidities could introduce bias [19]. Duarte et al. mentioned that the NASH group was composed only of female patients, which could have influenced the results [29]. The other studies did not report if there were any confounding factors, which is concerning since this is a common occurrence in clinical settings.
The methods used to detect and measure the Bifidobacterium genus in the microbiome can also offer insights into the reliability of the research papers. In particular, the NGS technology, the sequencing type, the reference database, and the statistical methods used to define the differential abundance of the genus among groups are of primary relevance.
Several studies on amplicon sequencing use the V3–V4 region, which is the standard in microbiome studies. However, some papers use other 16S regions, such as V3, V4–V5, and V5–V6, making it difficult to compare results across studies. The database is a significant concern in several studies, mainly because they rely on Greengenes, an ancient and outdated database last updated in 2013. On the other hand, the SILVA and the Ribosomal Database Project (RDP) continuously receive taxonomy revisions and updates. Furthermore, six studies did not identify which database they used, which is detrimental to research and its reproducibility.
Another important point concerns the statistical method used to assess the differences in bacterial abundance among different groups. Some studies used advanced methods such as LEfSe for microbiome analysis. In contrast, others relied on more straightforward statistics, such as the Krustal–Wallis or Wilcoxon test. The statistical methods used to analyze the microbiota need to be well-considered. A recent study demonstrated that using the Wilcoxon test on normalized data with a centered log-ratio led to the detection of up to 90% of bacteria in two sample groups. However, when employing Analysis of Compositions of Microbiomes (ANCOM)—a more robust, current, and suitable mathematical tool for microbiota analysis—bacterial detection decreased to 0.8% in the same sample data [76]. Other analytically more effective microbiome techniques than classical methods include LEfSe and Analysis of Compositions of Microbiomes with Bias Correction (ANCOM–BC). Among these, ANCOM–BC, being the most contemporary, offers additional advantages such as sampling fraction control, integrated statistical tests, confidence intervals, and considerable computational efficiency [77]. However, no study used the ANCOM–BC method. Furthermore, six studies omitted the statistical method utilized to assess variances in the Bifidobacterium genus, which is problematic.
It is necessary to reduce bias to ensure the quality of study results. This can be achieved through proper design and following validated checklists such as JBI. Additionally, choosing an updated database and using robust statistical methodologies for microbiota analysis should be considered to better understand the relationship between the Bifidobacterium genus and liver disease. On this aspect, new checklists have been proposed to improve reporting in future human microbiome studies, such as the “Strengthening The Organization and Reporting of Microbiome Studies” (STORMS) [78].
5. Conclusions
This comprehensive review has revealed a reduced abundance of Bifidobacterium in all stages of liver disease, such as ALD, NAFLD, NASH, cirrhosis, and HCC. However, interventional studies using different drugs and treatments were able to increase the abundance of the genus and improve clinical outcomes. By evaluating, in detail, the different studies, this review sheds light on the complex relationship between Bifidobacterium and liver diseases, as well as its progression in different stages. Various mechanisms are at play in this process, including the amplification of SCFA production, the dampening of inflammation, and the reinforcement of intestinal mucosal integrity. In addition, we have critically accessed the NGS methods and related statistical analyses of each study, highlighting concerns with the methods used to define the differential abundance of Bifidobacterium in some studies, including potential bias and the omission of relevant information.
Our better understanding of the underlying mechanisms of how this particular genus is related to liver injuries may help the development of new treatments that target these mechanisms. Furthermore, choosing an updated database and using robust statistical methodologies for microbiota analysis should be considered to better understand the relationship between the Bifidobacterium genus and liver disease.
References
- Pezzino, S.; Sofia, M.; Faletra, G.; Mazzone, C.; Litrico, G.; La Greca, G.; Latteri, S. Gut–liver Axis and Non-alcoholic Fatty Liver Disease: A Vicious Circle of Dysfunctions Orchestrated by the Gut Microbiome. Biology 2022, 11, 1622. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.P.; Suk, K.T.; Kim, D.J. Significance of Gut Microbiota in Alcoholic and Non-Alcoholic Fatty Liver Diseases. World J. Gastroenterol. 2021, 27, 6161–6179. [Google Scholar] [CrossRef] [PubMed]
- Seitz, H.K.; Bataller, R.; Cortez-Pinto, H.; Gao, B.; Gual, A.; Lackner, C.; Mathurin, P.; Mueller, S.; Szabo, G.; Tsukamoto, H. Alcoholic Liver Disease. Nat. Rev. Dis. Primers 2018, 4, 16. [Google Scholar] [CrossRef] [PubMed]
- Huo, R.; Chen, Y.; Li, J.; Xu, Q.; Guo, J.; Xu, H.; You, Y.; Zheng, C.; Chen, Y. Altered Gut Microbiota Composition and its Potential Association in Patients with Advanced Hepatocellular Carcinoma. Curr. Oncol. 2023, 30, 1818–1830. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, A.; Mandal, S. Bifidobacteria—Insight into Clinical Outcomes and Mechanisms of its Probiotic Action. Microbiol. Res. 2016, 192, 159–171. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Jiao, T.; Yu, Q.; Wang, J.; Wang, L.; Wang, G.; Zhang, H.; Zhao, J.; Chen, W. Bifidobacterium bifidum Shows More Diversified Ways of Relieving Non-Alcoholic Fatty Liver Compared with Bifidobacterium adolescentis. Biomedicines 2022, 10, 84. [Google Scholar] [CrossRef] [PubMed]
- Shahi, S.K.; Zarei, K.; Guseva, N.V.; Mangalam, A.K. Microbiota Analysis Using Two-step PCR and Next-generation 16S rRNA Gene Sequencing. J. Vis. Exp. 2019, 152, e59980. [Google Scholar] [CrossRef]
- Sanschagrin, S.; Yergeau, E. Next-generation Sequencing of 16S Ribosomal RNA Gene Amplicons. J. Vis. Exp. 2014, 90, e51709. [Google Scholar] [CrossRef]
- Ranjan, R.; Rani, A.; Metwally, A.; McGee, H.S.; Perkins, D.L. Analysis of the Microbiome: Advantages of Whole Genome Shotgun Versus 16S Amplicon Sequencing. Biochem. Biophys. Res. Commun. 2016, 469, 967–977. [Google Scholar] [CrossRef]
- Purushothaman, S.; Meola, M.; Egli, A. Combination of Whole Genome Sequencing and Metagenomics for Microbiological Diagnostics. Int. J. Mol. Sci. 2022, 23, 9834. [Google Scholar] [CrossRef]
- Marquardt, J.U.; Andersen, J.B. Next-generation Sequencing: Application in Liver Cancer—Past, Present and Future? Biology 2012, 1, 383–394. [Google Scholar] [CrossRef] [PubMed]
- Ouzzani, M.; Hammady, H.; Fedorowicz, Z.; Elmagarmid, A. Rayyan—A Web and Mobile App for Systematic Reviews. Syst. Rev. 2016, 5, 210. [Google Scholar] [CrossRef] [PubMed]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G.; PRISMA Group. Preferred Reporting Items for Systematic Reviews and Meta-Analyses: The PRISMA statement. PLoS Med. 2009, 6, e1000097. [Google Scholar] [CrossRef] [PubMed]
- Moola, S.; Munn, Z.; Tufanaru, C.; Aromataris, E.; Sears, K.; Sfetcu, R.; Currie, M.; Lisy, K.; Qureshi, R.; Mattis, P.; et al. Chapter 7: Systematic Reviews of Etiology and Risk. In JBI Manual for Evidence Synthesis; Aromataris, E., Munn, Z., Eds.; JBI: Adelaide, Australia, 2020. [Google Scholar] [CrossRef]
- Gu, Z.; Wu, Y.; Wang, Y.; Sun, H.; You, Y.; Piao, C.; Liu, J.; Wang, Y. Lactobacillus rhamnosus Granules Dose-Dependently Balance Intestinal Microbiome Disorders and Ameliorate Chronic Alcohol-Induced Liver Injury. J. Med. Food 2020, 23, 114–124. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Coker, O.O.; Chu, E.S.; Fu, K.; Lau, H.C.H.; Wang, Y.-X.; Chan, A.W.H.; Wei, H.; Yang, X.; Sung, J.J.Y.; et al. Dietary Cholesterol Drives Fatty Liver-Associated Liver Cancer by Modulating Gut Microbiota and Metabolites. Gut 2021, 70, 761–774. [Google Scholar] [CrossRef] [PubMed]
- Ponziani, F.R.; Bhoori, S.; Castelli, C.; Putignani, L.; Rivoltini, L.; Del Chierico, F.; Sanguinetti, M.; Morelli, D.; Sterbini, F.P.; Petito, V.; et al. Hepatocellular Carcinoma is Associated with Gut Microbiota Profile and Inflammation in Nonalcoholic Fatty Liver Disease. Hepatology 2019, 69, 107–120. [Google Scholar] [CrossRef] [PubMed]
- Kajihara, M.; Koido, S.; Kanai, T.; Ito, Z.; Matsumoto, Y.; Takakura, K.; Saruta, M.; Kato, K.; Odamaki, T.; Xiao, J.-Z.; et al. Characterisation of Blood Microbiota in Patients with Liver Cirrhosis. Eur. J. Gastroenterol. Hepatol. 2019, 31, 1577–1583. [Google Scholar] [CrossRef]
- Wei, X.; Yan, X.; Zou, D.; Yang, Z.; Wang, X.; Liu, W.; Wang, S.; Li, X.; Han, J.; Huang, L.; et al. Abnormal Fecal Microbiota Community and Functions in Patients with Hepatitis B Liver Cirrhosis as Revealed by a Metagenomic Approach. BMC Gastroenterol. 2013, 13, 175. [Google Scholar] [CrossRef]
- Dubinkina, V.B.; Tyakht, A.V.; Odintsova, V.Y.; Yarygin, K.S.; Kovarsky, B.A.; Pavlenko, A.V.; Ischenko, D.S.; Popenko, A.S.; Alexeev, D.G.; Taraskina, A.Y.; et al. Links of Gut Microbiota Composition with Alcohol Dependence Syndrome and Alcoholic Liver Disease. Microbiome 2017, 5, 141. [Google Scholar] [CrossRef]
- Juárez-Fernández, M.; Porras, D.; Petrov, P.; Román-Sagüillo, S.; García-Mediavilla, M.V.; Soluyanova, P.; Martínez-Flórez, S.; González-Gallego, J.; Nistal, E.; Jover, R.; et al. The Synbiotic Combination of Akkermansia muciniphila and Quercetin Ameliorates Early Obesity and NAFLD through Gut Microbiota Reshaping and Bile Acid Metabolism Modulation. Antioxidants 2021, 10, 2001. [Google Scholar] [CrossRef]
- Zhang, Z.; Chen, X.; Cui, B. Modulation of the Fecal Microbiome and Metabolome by Resistant Dextrin Ameliorates Hepatic Steatosis and Mitochondrial Abnormalities in Mice. Food Funct. 2021, 12, 4504–4518. [Google Scholar] [CrossRef] [PubMed]
- Kang, K.; Sun, Y.; Pan, D.; Sang, L.-X.; Sun, M.-J.; Li, Y.-L.; Chang, B. Distinctive Gut Microbial Dysbiosis Between Chronic Alcoholic Fatty Liver Disease and Metabolic-Associated Fatty Liver Disease in Mice. Exp. Ther. Med. 2021, 21, 418. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Sun, J.; Yu, J.; Chen, H.; Zhang, D.; Zhang, T.; Ma, Y.; Zou, C.; Zhang, Z.; Ma, L.; et al. Gut Microbiome Determines Therapeutic Effects of OCA on NAFLD by Modulating Bile Acid Metabolism. npj Biofilms Microbiomes 2023, 9, 29. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Zhang, K.; Yang, H. Pectin Alleviates High Fat (Lard) Diet-Induced Nonalcoholic Fatty Liver Disease in Mice: Possible Role of Short-Chain Fatty Acids and Gut Microbiota Regulated by Pectin. J. Agric. Food Chem. 2018, 66, 8015–8025. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Luo, X.; Han, C.; Liu, J.; Zhang, L.; Qi, J.; Gu, J.; Tan, R.; Gong, P. Terminalia bellirica Ethanol Extract Ameliorates Nonalcoholic Fatty Liver Disease in Mice by Amending the Intestinal Microbiota and Faecal Metabolites. J. Ethnopharmacol. 2023, 305, 116082. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.-Z.; Li, Y.-T.; Wu, W.-R.; Shi, D.; Fang, D.-Q.; Yang, L.-Y.; Bian, X.-Y.; Wu, J.-J.; Wang, Q.; Jiang, X.-W.; et al. Dynamic Alterations in the Gut Microbiota and Metabolome During the Development of Methionine-Choline-Deficient Diet-Induced Nonalcoholic Steatohepatitis. World J. Gastroenterol. 2018, 24, 2468–2481. [Google Scholar] [CrossRef] [PubMed]
- Nobili, V.; Putignani, L.; Mosca, A.; Del Chierico, F.; Vernocchi, P.; Alisi, A.; Stronati, L.; Cucchiara, S.; Toscano, M.; Drago, L. Bifidobacteria and Lactobacilli in the Gut Microbiome of Children with Non-Alcoholic Fatty Liver Disease: Which Strains Act as Health Players? Arch. Med. Sci. 2018, 14, 81–87. [Google Scholar] [CrossRef]
- Duarte, S.M.B.; Stefano, J.T.; Miele, L.; Ponziani, F.R.; Souza-Basqueira, M.; Okada, L.S.R.R.; de Barros Costa, F.G.; Toda, K.; Mazo, D.F.C.; Sabino, E.C.; et al. Gut Microbiome Composition in Lean Patients with NASH is Associated with Liver Damage Independent of Caloric Intake: A Prospective Pilot Study. Nutr. Metab. Cardiovasc. Dis. 2018, 28, 369–384. [Google Scholar] [CrossRef]
- Pan, X.; Kaminga, A.C.; Liu, A.; Wen, S.W.; Luo, M.; Luo, J. Gut Microbiota, Glucose, Lipid, and Water-Electrolyte Metabolism in Children with Nonalcoholic Fatty Liver Disease. Front. Cell. Infect. Microbiol. 2021, 11, 683743. [Google Scholar] [CrossRef]
- Warner, J.B.; Larsen, I.S.; Hardesty, J.E.; Song, Y.L.; Warner, D.R.; McClain, C.J.; Sun, R.; Deng, Z.; Jensen, B.A.H.; Kirpich, I.A. Human Beta Defensin 2 Ameliorated Alcohol-Associated Liver Disease in Mice. Front. Physiol. 2022, 12, 812882. [Google Scholar] [CrossRef]
- Lin, Y.; Chen, H.; Cao, Y.; Zhang, Y.; Li, W.; Guo, W.; Lv, X.; Rao, P.; Ni, L.; Liu, P. Auricularia auricula Melanin Protects against Alcoholic Liver Injury and Modulates Intestinal Microbiota Composition in Mice Exposed to Alcohol Intake. Foods 2021, 10, 2436. [Google Scholar] [CrossRef] [PubMed]
- Philips, C.A.; Ahamed, R.; Rajesh, S.; Singh, S.; Tharakan, A.; Abduljaleel, J.K.; Augustine, P. Clinical Outcomes and Gut Microbiota Analysis of Severe Alcohol-Associated Hepatitis Patients Undergoing Healthy Donor Fecal Transplant or Pentoxifylline Therapy: Single-Center Experience from Kerala. Gastroenterol. Rep. 2022, 10, goac074. [Google Scholar] [CrossRef] [PubMed]
- Ponziani, F.R.; De Luca, A.; Picca, A.; Marzetti, E.; Petito, V.; Del Chierico, F.; Reddel, S.; Sterbini, F.P.; Sanguinetti, M.; Putignani, L.; et al. Gut Dysbiosis and Fecal Calprotectin Predict Response to Immune Checkpoint Inhibitors in Patients with Hepatocellular Carcinoma. Hepatol. Commun. 2022, 6, 1492–1501. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Chen, L.; Pan, X.; Yao, Y.; Zhang, H.; Zhu, X.; Lou, X.; Zhu, C.; Wang, J.; Li, L.; et al. Lactitol Supplementation Modulates Intestinal Microbiome in Liver Cirrhotic Patients. Front. Med. 2021, 8, 762930. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Tang, K.; Deng, Y.; Chen, R.; Liang, S.; Xie, H.; He, Y.; Chen, Y.; Yang, Q. Effects of Shenling Baizhu Powder Herbal Formula on Intestinal Microbiota in High-Fat Diet-Induced NAFLD Rats. Biomed. Pharmacother. 2018, 102, 1025–1036. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Feng, B.; Niu, Y.J.; Hu, C.Y.; Meng, Y.H. Fu Instant Tea Ameliorates Fatty Liver by Improving Microbiota Dysbiosis and Elevating Short-Chain Fatty Acids in the Intestine of Mice Fed a High-Fat Diet. Food Biosci. 2021, 42, 101207. [Google Scholar] [CrossRef]
- Bao, T.; He, F.; Zhang, X.; Zhu, L.; Wang, Z.; Lu, H.; Wang, T.; Li, Y.; Yang, S.; Wang, H. Inulin Exerts Beneficial Effects on Non-Alcoholic Fatty Liver Disease via Modulating gut Microbiome and Suppressing the Lipopolysaccharide-Toll-Like Receptor 4-Mψ-Nuclear Factor-κB-Nod-Like Receptor Protein 3 Pathway via Gut-Liver Axis in Mice. Front. Pharmacol. 2020, 11, 558525. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Su, H.; Lv, Y.; Tao, H.; Jiang, Y.; Ni, Z.; Peng, L.; Chen, X. Inulin Intervention Attenuates Hepatic Steatosis in Rats Via Modulating Gut Microbiota and Maintaining Intestinal Barrier Function. Food Res. Int. 2023, 163, 112309. [Google Scholar] [CrossRef]
- Hu, W.; Gao, W.; Liu, Z.; Fang, Z.; Wang, H.; Zhao, J.; Zhang, H.; Lu, W.; Chen, W. Specific Strains of Faecalibacterium prausnitzii Ameliorate Nonalcoholic Fatty Liver Disease in Mice in Association with Gut Microbiota Regulation. Nutrients 2022, 14, 2945. [Google Scholar] [CrossRef]
- Ghosh, S.; Yang, X.; Wang, L.; Zhang, C.; Zhao, L. Active Phase Prebiotic Feeding Alters Gut Microbiota, Induces Weight-Independent Alleviation of Hepatic Steatosis and Serum Cholesterol in High-Fat Diet-Fed Mice. Comput. Struct. Biotechnol. J. 2021, 19, 448–458. [Google Scholar] [CrossRef]
- Abernathy, B.E.; Schoenfuss, T.C.; Bailey, A.S.; Gallaher, D.D. Polylactose Exhibits Prebiotic Activity and Reduces Adiposity and Nonalcoholic Fatty Liver Disease in Rats Fed a High-Fat Diet. J. Nutr. 2021, 151, 352–360. [Google Scholar] [CrossRef] [PubMed]
- Satapathy, S.K.M.; Banerjee, P.; Pierre, J.F.; Higgins, D.; Dutta, S.; Heda, R.B.; Khan, S.D.B.; Mupparaju, V.K.; Mas, V.; Nair, S.; et al. Characterization of Gut Microbiome in Liver Transplant Recipients with Nonalcoholic Steatohepatitis. Transplant. Direct 2020, 6, e625. [Google Scholar] [CrossRef] [PubMed]
- Bomhof, M.R.; Parnell, J.A.; Ramay, H.R.; Crotty, P.; Rioux, K.P.; Probert, C.S.; Jayakumar, S.; Raman, M.; Reimer, R.A. Histological Improvement of Non-Alcoholic Steatohepatitis with a Prebiotic: A Pilot Clinical Trial. Eur. J. Nutr. 2019, 58, 1735–1745. [Google Scholar] [CrossRef] [PubMed]
- Bessone, F.; Razori, M.V.; Roma, M.G. Molecular Pathways of Nonalcoholic Fatty Liver Disease Development and Progression. Cell. Mol. Life Sci. 2019, 76, 99–128. [Google Scholar] [CrossRef] [PubMed]
- Ceni, E.; Mello, T.; Galli, A. Pathogenesis of Alcoholic Liver Disease: Role of Oxidative Metabolism. World J. Gastroenterol. 2014, 20, 17756–17772. [Google Scholar] [CrossRef] [PubMed]
- Dolicka, D.; Sobolewski, C.; Correia de Sousa, M.; Gjorgjieva, M.; Foti, M. mRNA Post-Transcriptional Regulation by AU-Rich Element-Binding Proteins in Liver Inflammation and Cancer. Int. J. Mol. Sci. 2020, 21, 6648. [Google Scholar] [CrossRef] [PubMed]
- Sgorbati, B.; Biavati, B.; Palenzona, D. The Genus Bifidobacterium. In The Genera of Lactic Acid Bacteria; Wood, B.J.B., Holzapfel, W.H., Eds.; Springer: New York, NY, USA, 1995; Volume 2. [Google Scholar] [CrossRef]
- Parte, A.C. LPSN—List of Prokaryotic Names with Standing in Nomenclature. Nucleic Acids Res. 2014, 42, D613–D616. [Google Scholar] [CrossRef] [PubMed]
- Turroni, F.; van Sinderen, D.; Ventura, M. Genomics and Ecological Overview of the Genus Bifidobacterium. Int. J. Food Microbiol. 2011, 149, 37–44. [Google Scholar] [CrossRef]
- Arboleya, S.; Watkins, C.; Stanton, C.; Ross, R.P. Gut Bifidobacteria Populations in Human Health and Aging. Front. Microbiol. 2016, 7, 1204. [Google Scholar] [CrossRef]
- Zhang, J.; Sun, Z.; Jiang, S.; Bai, X.; Ma, C.; Peng, Q.; Chen, K.; Chang, H.; Fang, T.; Zhang, H. Probiotic Bifidobacterium lactis V9 Regulates the Secretion of Sex Hormones in Polycystic Ovary Syndrome Patients through the Gut-Brain Axis. mSystems 2019, 4, e00017-19. [Google Scholar] [CrossRef]
- Lim, H.J.; Shin, H.S. Antimicrobial and Immunomodulatory Effects of Bifidobacterium Strains: A Review. J. Microbiol. Biotechnol. 2020, 30, 1793–1800. [Google Scholar] [CrossRef]
- Gurung, M.; Li, Z.; You, H.; Rodrigues, R.; Jump, D.B.; Morgun, A.; Shulzhenko, N. Role of Gut Microbiota in Type 2 Diabetes Pathophysiology. eBioMedicine 2020, 51, 102590. [Google Scholar] [CrossRef] [PubMed]
- Cheung, S.G.; Goldenthal, A.R.; Uhlemann, A.C.; Mann, J.J.; Miller, J.M.; Sublette, M.E. Systematic Review of Gut Microbiota and Major Depression. Front. Psychiatry 2019, 10, 34. [Google Scholar] [CrossRef] [PubMed]
- Liwinski, T.; Casar, C.; Ruehlemann, M.C.; Bang, C.; Sebode, M.; Hohenester, S.; Denk, G.; Lieb, W.; Lohse, A.W.; Franke, A.; et al. A Disease-Specific Decline of the Relative Abundance of Bifidobacterium in Patients with Autoimmune Hepatitis. Aliment. Pharmacol. Ther. 2020, 51, 1417–1428. [Google Scholar] [CrossRef] [PubMed]
- Bellentani, S.; Tiribelli, C. The Spectrum of Liver Disease in the General Population: Lesson from the Dionysos Study. J. Hepatol. 2001, 35, 531–537. [Google Scholar] [CrossRef] [PubMed]
- Gurwara, S.; Dai, A.; Ajami, N.J.; Graham, D.Y.; White, D.L.; Chen, L.; Jang, A.; Chen, E.; El-Serag, H.B.; Petrosino, J.F.; et al. Alcohol Use Alters the Colonic Mucosa–Associated Gut Microbiota in Humans. Nutr. Res. 2020, 83, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Kolodziejczyk, A.A.; Zheng, D.; Shibolet, O.; Elinav, E. The Role of the Microbiome in NAFLD and NASH. EMBO Mol. Med. 2019, 11, e9302. [Google Scholar] [CrossRef] [PubMed]
- Velázquez, K.T.; Enos, R.T.; E Bader, J.; Sougiannis, A.T.; Carson, M.S.; Chatzistamou, I.; A Carson, J.; Nagarkatti, P.; Nagarkatti, M.; Murphy, E.A. Prolonged High-Fat-Diet Feeding Promotes Non-Alcoholic Fatty Liver Disease and Alters Gut Microbiota in Mice. World J. Hepatol. 2019, 11, 619–637. [Google Scholar] [CrossRef] [PubMed]
- Leung, C.; Rivera, L.; Furness, J.B.; Angus, P.W. The Role of the Gut Microbiota in NAFLD. Nat. Rev. Gastroenterol. Hepatol. 2016, 13, 412–425. [Google Scholar] [CrossRef]
- McBurney, M.I.; Davis, C.; Fraser, C.M.; Schneeman, B.O.; Huttenhower, C.; Verbeke, K.; Walter, J.; Latulippe, M.E. Establishing What Constitutes a Healthy Human Gut Microbiome: State of the Science, Regulatory Considerations, and Future Directions. J. Nutr. 2019, 149, 1882–1895. [Google Scholar] [CrossRef]
- Li, F.; Ye, J.; Shao, C.; Zhong, B. Compositional Alterations of Gut Microbiota in Nonalcoholic Fatty Liver Disease Patients: A Systematic Review and Meta-analysis. Lipids Health Dis. 2021, 20, 22. [Google Scholar] [CrossRef] [PubMed]
- Honda, K.; Littman, D. The Microbiota in Adaptive Immune Homeostasis and Disease. Nature 2016, 535, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Jokela, R.; Ponsero, A.J.; Dikareva, E.; Wei, X.; Kolho, K.L.; Korpela, K.; de Vos, W.M.; Salonen, A. Sources of Gut Microbiota Variation in a Large Longitudinal Finnish Infant Cohort. EBioMedicine. 2023, 94, 104695. [Google Scholar] [CrossRef] [PubMed]
- Sheng, W.; Ji, G.; Zhang, L. Immunomodulatory Effects of Inulin and its Intestinal Metabolites. Front Immunol. 2023, 14, 1224092. [Google Scholar] [CrossRef] [PubMed]
- McCullough, A.J. Update on Nonalcoholic Fatty Liver Disease. J. Clin. Gastroenterol. 2002, 34, 255–262. [Google Scholar] [CrossRef] [PubMed]
- Tsay, C.J.; Lim, J.K. NASH and the Gut Microbiome: Implications for New Therapies. Clin. Liver Dis. 2022, 19, 97–100. [Google Scholar] [CrossRef] [PubMed]
- Schuppan, D.; Afdhal, N.H. Liver Cirrhosis. Lancet 2008, 371, 838–851. [Google Scholar] [CrossRef] [PubMed]
- Berzigotti, A. Advances and Challenges in Cirrhosis and Portal Hypertension. BMC Med. 2017, 15, 200. [Google Scholar] [CrossRef]
- Cervantes-Alvarez, E.; Vilatoba, M.; la Rosa, N.L.-D.; Mendez-Guerrero, O.; Kershenobich, D.; Torre, A.; Navarro-Alvarez, N. Liver Transplantation is Beneficial Regardless of Cirrhosis Stage or Acute-on-Chronic Liver Failure Grade: A Single-Center Experience. World J. Gastroenterol. 2022, 28, 5881–5892. [Google Scholar] [CrossRef]
- Bajaj, J.S. Altered Microbiota in Cirrhosis and its Relationship to the Development of Infection. Clin. Liver Dis. 2019, 14, 107–111. [Google Scholar] [CrossRef]
- Ganesan, P.; Kulik, L.M. Hepatocellular Carcinoma: New Developments. Clin. Liver Dis. 2023, 27, 85–102. [Google Scholar] [CrossRef] [PubMed]
- Dhanasekaran, R.; Bandoh, S.; Roberts, L.R. Molecular Pathogenesis of Hepatocellular Carcinoma and Impact of Therapeutic Advances. F1000Research 2016, 5, 879. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; Guo, S.; Zhou, Y.; Zhao, J.; Wang, M.; Sang, L.; Chang, B.; Wang, B. Hepatocellular Carcinoma: How the Gut Microbiota Contributes to Pathogenesis, Diagnosis, and Therapy. Front. Microbiol. 2022, 13, 873160. [Google Scholar] [CrossRef] [PubMed]
- Litwinowicz, K.; Gamian, A. Microbiome Alterations in Alcohol Use Disorder and Alcoholic Liver Disease. Int. J. Mol. Sci. 2023, 24, 2461. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Peddada, S.D. Analysis of Compositions of Microbiomes with Bias Correction. Nat. Commun. 2020, 11, 3514. [Google Scholar] [CrossRef] [PubMed]
- Mirzayi, C.; Renson, A.; Genomic Standards Consortium; Massive Analysis and Quality Control Society; Zohra, F.; Elsafoury, S.; Geistlinger, L.; Kasselman, L.J.; Eckenrode, K.; van de Wijgert, J.; et al. Reporting Guidelines for Human Microbiome Research: The STORMS Checklist. Nat. Med. 2021, 27, 1885–1892. [Google Scholar] [CrossRef] [PubMed]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef]

